Functional cis-regulatory genomics for systems biology
- PMID: 20142491
- PMCID: PMC2840491
- DOI: 10.1073/pnas.1000147107
Functional cis-regulatory genomics for systems biology
Abstract
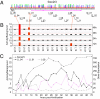
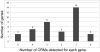
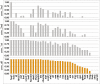
Gene expression is controlled by interactions between trans-regulatory factors and cis-regulatory DNA sequences, and these interactions constitute the essential functional linkages of gene regulatory networks (GRNs). Validation of GRN models requires experimental cis-regulatory tests of predicted linkages to authenticate their identities and proposed functions. However, cis-regulatory analysis is, at present, at a severe bottleneck in genomic system biology because of the demanding experimental methodologies currently in use for discovering cis-regulatory modules (CRMs), in the genome, and for measuring their activities. Here we demonstrate a high-throughput approach to both discovery and quantitative characterization of CRMs. The unique aspect is use of DNA sequence tags to "barcode" CRM expression constructs, which can then be mixed, injected together into sea urchin eggs, and subsequently deconvolved. This method has increased the rate of cis-regulatory analysis by >100-fold compared with conventional one-by-one reporter assays. The utility of the DNA-tag reporters was demonstrated by the rapid discovery of 81 active CRMs from 37 previously unexplored sea urchin genes. We then obtained simultaneous high-resolution temporal characterization of the regulatory activities of more than 80 CRMs. On average 2-3 CRMs were discovered per gene. Comparison of endogenous gene expression profiles with those of the CRMs recovered from each gene showed that, for most cases, at least one CRM is active in each phase of endogenous expression, suggesting that CRM recovery was comprehensive. This approach will qualitatively alter the practice of GRN construction as well as validation, and will impact many additional areas of regulatory system biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Single embryo-resolution quantitative analysis of reporters permits multiplex spatial cis-regulatory analysis.Dev Biol. 2017 Feb 15;422(2):92-104. doi: 10.1016/j.ydbio.2017.01.010. Epub 2017 Jan 16. Dev Biol. 2017. PMID: 28099870
-
Barcoded DNA-tag reporters for multiplex cis-regulatory analysis.PLoS One. 2012;7(4):e35934. doi: 10.1371/journal.pone.0035934. Epub 2012 Apr 26. PLoS One. 2012. PMID: 22563420 Free PMC article.
-
A systems biology approach to understanding cis-regulatory module function.Semin Cell Dev Biol. 2009 Sep;20(7):856-62. doi: 10.1016/j.semcdb.2009.07.007. Epub 2009 Aug 4. Semin Cell Dev Biol. 2009. PMID: 19660565 Review.
-
Global analysis of primary mesenchyme cell cis-regulatory modules by chromatin accessibility profiling.BMC Genomics. 2018 Mar 20;19(1):206. doi: 10.1186/s12864-018-4542-z. BMC Genomics. 2018. PMID: 29558892 Free PMC article.
-
Enhancer evolution in chordates: Lessons from functional analyses of cephalochordate cis-regulatory modules.Dev Growth Differ. 2020 Jun;62(5):279-300. doi: 10.1111/dgd.12684. Epub 2020 Jun 16. Dev Growth Differ. 2020. PMID: 32479656 Review.
Cited by
-
Gene regulatory control in the sea urchin aboral ectoderm: spatial initiation, signaling inputs, and cell fate lockdown.Dev Biol. 2013 Feb 1;374(1):245-54. doi: 10.1016/j.ydbio.2012.11.013. Epub 2012 Dec 2. Dev Biol. 2013. PMID: 23211652 Free PMC article.
-
Function-based identification of mammalian enhancers using site-specific integration.Nat Methods. 2014 May;11(5):566-71. doi: 10.1038/nmeth.2886. Epub 2014 Mar 23. Nat Methods. 2014. PMID: 24658141 Free PMC article.
-
The analysis of novel distal Cebpa enhancers and silencers using a transcriptional model reveals the complex regulatory logic of hematopoietic lineage specification.Dev Biol. 2016 May 1;413(1):128-44. doi: 10.1016/j.ydbio.2016.02.030. Epub 2016 Mar 3. Dev Biol. 2016. PMID: 26945717 Free PMC article.
-
Bacterial artificial chromosomes as recombinant reporter constructs to investigate gene expression and regulation in echinoderms.Brief Funct Genomics. 2018 Sep 27;17(5):362-371. doi: 10.1093/bfgp/elx031. Brief Funct Genomics. 2018. PMID: 29045542 Free PMC article. Review.
-
Transcription factor NFYa controls cardiomyocyte metabolism and proliferation during mouse fetal heart development.Dev Cell. 2023 Dec 18;58(24):2867-2880.e7. doi: 10.1016/j.devcel.2023.10.012. Epub 2023 Nov 15. Dev Cell. 2023. PMID: 37972593 Free PMC article.
References
-
- Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. San Diego: Academic Press/Elsevier; 2006.
-
- Davidson EH, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

