The DNA binding CXC domain of MSL2 is required for faithful targeting the Dosage Compensation Complex to the X chromosome
- PMID: 20139418
- PMCID: PMC2879509
- DOI: 10.1093/nar/gkq026
The DNA binding CXC domain of MSL2 is required for faithful targeting the Dosage Compensation Complex to the X chromosome
Abstract
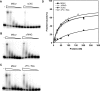
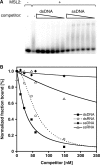
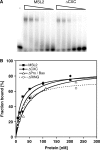
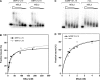
Dosage compensation in Drosophila melanogaster involves the selective targeting of the male X chromosome by the dosage compensation complex (DCC) and the coordinate, approximately 2-fold activation of most genes. The principles that allow the DCC to distinguish the X chromosome from the autosomes are not understood. Targeting presumably involves DNA sequence elements whose combination or enrichment mark the X chromosome. DNA sequences that characterize 'chromosomal entry sites' or 'high-affinity sites' may serve such a function. However, to date no DNA binding domain that could interpret sequence information has been identified within the subunits of the DCC. Early genetic studies suggested that MSL1 and MSL2 serve to recognize high-affinity sites (HAS) in vivo, but a direct interaction of these DCC subunits with DNA has not been studied. We now show that recombinant MSL2, through its CXC domain, directly binds DNA with low nanomolar affinity. The DNA binding of MSL2 or of an MSL2-MSL1 complex does not discriminate between different sequences in vitro, but in a reporter gene assay in vivo, suggesting the existence of an unknown selectivity cofactor. Reporter gene assays and localization of GFP-fusion proteins confirm the important contribution of the CXC domain for DCC targeting in vivo.
Figures



Similar articles
-
Msl1-mediated dimerization of the dosage compensation complex is essential for male X-chromosome regulation in Drosophila.Mol Cell. 2012 Nov 30;48(4):587-600. doi: 10.1016/j.molcel.2012.09.014. Epub 2012 Oct 18. Mol Cell. 2012. PMID: 23084835
-
Structural basis of X chromosome DNA recognition by the MSL2 CXC domain during Drosophila dosage compensation.Genes Dev. 2014 Dec 1;28(23):2652-62. doi: 10.1101/gad.250936.114. Genes Dev. 2014. PMID: 25452275 Free PMC article.
-
PionX sites mark the X chromosome for dosage compensation.Nature. 2016 Sep 8;537(7619):244-248. doi: 10.1038/nature19338. Epub 2016 Aug 31. Nature. 2016. PMID: 27580037
-
Chromatin mechanisms in Drosophila dosage compensation.Prog Mol Subcell Biol. 2005;38:123-49. doi: 10.1007/3-540-27310-7_5. Prog Mol Subcell Biol. 2005. PMID: 15881893 Review.
-
Dosage Compensation of the X Chromosome: A Complex Epigenetic Assignment Involving Chromatin Regulators and Long Noncoding RNAs.Annu Rev Biochem. 2018 Jun 20;87:323-350. doi: 10.1146/annurev-biochem-062917-011816. Epub 2018 Apr 18. Annu Rev Biochem. 2018. PMID: 29668306 Review.
Cited by
-
The CLAMP protein links the MSL complex to the X chromosome during Drosophila dosage compensation.Genes Dev. 2013 Jul 15;27(14):1551-6. doi: 10.1101/gad.214585.113. Genes Dev. 2013. PMID: 23873939 Free PMC article.
-
The Drosophila over compensating males gene genetically inhibits dosage compensation in males.PLoS One. 2013;8(4):e60450. doi: 10.1371/journal.pone.0060450. Epub 2013 Apr 2. PLoS One. 2013. PMID: 23565249 Free PMC article.
-
Physical interaction between MSL2 and CLAMP assures direct cooperativity and prevents competition at composite binding sites.Nucleic Acids Res. 2023 Sep 22;51(17):9039-9054. doi: 10.1093/nar/gkad680. Nucleic Acids Res. 2023. PMID: 37602401 Free PMC article.
-
The Drosophila CLAMP protein associates with diverse proteins on chromatin.PLoS One. 2017 Dec 27;12(12):e0189772. doi: 10.1371/journal.pone.0189772. eCollection 2017. PLoS One. 2017. PMID: 29281702 Free PMC article.
-
Divergent evolution toward sex chromosome-specific gene regulation in Drosophila.Genes Dev. 2021 Jul 1;35(13-14):1055-1070. doi: 10.1101/gad.348411.121. Epub 2021 Jun 17. Genes Dev. 2021. PMID: 34140353 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

