Engineering an artificial zymogen by alternate frame protein folding
- PMID: 20133757
- PMCID: PMC2840272
- DOI: 10.1073/pnas.0907668107
Engineering an artificial zymogen by alternate frame protein folding
Abstract
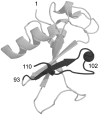
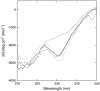
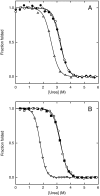
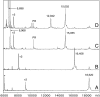
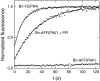
Alternate frame folding (AFF) is a novel mechanism by which allostery can be introduced into a protein where none may have existed previously. We employ this technology to convert the cytotoxic ribonuclease barnase into an artificial zymogen that is activated by HIV-1 protease. The AFF modification entails partial duplication of the polypeptide chain and mutation of a key catalytic residue in one of the duplicated segments. The resulting molecule can fold in one of two "frames" to yield the wild-type structure or a circularly permuted form in which the positions of the N- and C-termini are exchanged with a surface loop. It cannot take on both structures simultaneously because each competes for a shared amino acid sequence. An HIV-1 protease recognition sequence is inserted into one of the surface loops in the nonpermuted frame, and cleavage induces a shift from the nonpermuted fold to the permuted fold. Using the AFF mechanism, we were able to suppress k(cat)/K(M) by 250-fold in the proenzyme relative to wild-type barnase. HIV-1 protease cleavage subsequently increases k(cat)/K(M) by 130-fold. AFF is significant because it is general and can in principle be used to control activity of many enzymes, including those whose functions are not regulated by any existing mechanism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Circular zymogens of human ribonuclease 1.Protein Sci. 2019 Sep;28(9):1713-1719. doi: 10.1002/pro.3686. Epub 2019 Aug 6. Protein Sci. 2019. PMID: 31306518 Free PMC article.
-
Structural and thermodynamic analysis of a conformationally strained circular permutant of barnase.Biochemistry. 2009 Apr 21;48(15):3497-507. doi: 10.1021/bi900039e. Biochemistry. 2009. PMID: 19260676 Free PMC article.
-
(1)H, (13)C and (15)N resonance assignments of the Onconase FL-G zymogen.Biomol NMR Assign. 2013 Apr;7(1):13-5. doi: 10.1007/s12104-012-9367-0. Epub 2012 Mar 4. Biomol NMR Assign. 2013. PMID: 22392335
-
A new approach for alteration of protease functions: pro-sequence engineering.Appl Microbiol Biotechnol. 2003 Nov;63(1):1-9. doi: 10.1007/s00253-003-1352-1. Epub 2003 Jul 15. Appl Microbiol Biotechnol. 2003. PMID: 12879301 Review.
-
The folding of an enzyme. III. Structure of the transition state for unfolding of barnase analysed by a protein engineering procedure.J Mol Biol. 1992 Apr 5;224(3):805-18. doi: 10.1016/0022-2836(92)90563-y. J Mol Biol. 1992. PMID: 1569558 Review.
Cited by
-
Regulated unfolding of proteins in signaling.FEBS Lett. 2013 Apr 17;587(8):1081-8. doi: 10.1016/j.febslet.2013.02.024. Epub 2013 Feb 20. FEBS Lett. 2013. PMID: 23454209 Free PMC article. Review.
-
GFP variants with alternative β-strands and their application as light-driven protease sensors: a tale of two tails.J Am Chem Soc. 2013 Jul 17;135(28):10226-9. doi: 10.1021/ja4037274. Epub 2013 Jul 8. J Am Chem Soc. 2013. PMID: 23819615 Free PMC article.
-
Converting a protein into a switch for biosensing and functional regulation.Protein Sci. 2011 Jan;20(1):19-29. doi: 10.1002/pro.541. Protein Sci. 2011. PMID: 21064163 Free PMC article. Review.
-
Structural characterization of two alternate conformations in a calbindin D₉k-based molecular switch.Biochemistry. 2011 Jun 28;50(25):5583-9. doi: 10.1021/bi102040g. Epub 2011 Jun 1. Biochemistry. 2011. PMID: 21618991 Free PMC article.
-
Molecular simulations of mutually exclusive folding in a two-domain protein switch.Biophys J. 2011 Feb 2;100(3):756-764. doi: 10.1016/j.bpj.2010.12.3710. Biophys J. 2011. PMID: 21281591 Free PMC article.
References
-
- Johnson RJ, Lin SR, Raines RT. A ribonuclease zymogen activated by the NS3 protease of the hepatitis C virus. FEBS J. 2006;273:5457–5465. - PubMed
-
- Mossakowska DE, Nyberg K, Fersht AR. Kinetic characterization of the recombinant ribonuclease from Bacillus amyloliquifaciens (barnase) and investigation of key residues in catalysis by site-directed mutagenesis. Biochemistry. 1989;28:3843–3850. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

