High resolution electron transfer dissociation studies of unfractionated intact histones from murine embryonic stem cells using on-line capillary LC separation: determination of abundant histone isoforms and post-translational modifications
- PMID: 20133344
- PMCID: PMC2871417
- DOI: 10.1074/mcp.M900569-MCP200
High resolution electron transfer dissociation studies of unfractionated intact histones from murine embryonic stem cells using on-line capillary LC separation: determination of abundant histone isoforms and post-translational modifications
Abstract
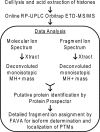
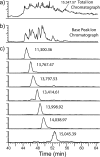
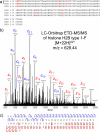
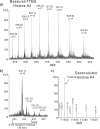
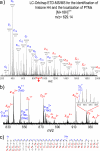
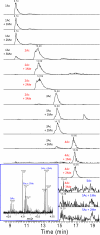
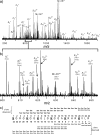
Epigenetic regulation of chromatin is dependent on both the histone protein isoforms and state of their post-translational modifications. The assignment of all post-translational modification sites for each individual intact protein isoform remains an experimental challenge. We present an on-line reversed phase LC tandem mass spectrometry approach for the separation of intact, unfractionated histones and a high resolution mass analyzer, the Orbitrap, with electron transfer dissociation capabilities to detect and record accurate mass values for the molecular and fragment ions observed. From a single LC-electron transfer dissociation run, this strategy permits the identification of the most abundant intact proteins, determination of the isoforms present, and the localization of post-translational modifications.
Figures
Similar articles
-
Quantitative mass spectrometry of histones H3.2 and H3.3 in Suz12-deficient mouse embryonic stem cells reveals distinct, dynamic post-translational modifications at Lys-27 and Lys-36.Mol Cell Proteomics. 2010 May;9(5):838-50. doi: 10.1074/mcp.M900489-MCP200. Epub 2010 Feb 11. Mol Cell Proteomics. 2010. PMID: 20150217 Free PMC article.
-
Data-dependent middle-down nano-liquid chromatography-electron capture dissociation-tandem mass spectrometry: an application for the analysis of unfractionated histones.Anal Chem. 2013 Apr 2;85(7):3501-7. doi: 10.1021/ac303103b. Epub 2013 Mar 12. Anal Chem. 2013. PMID: 23448339
-
Characterization of post-translational modifications of the linker histones H1 and H5 from chicken erythrocytes using mass spectrometry.J Proteome Res. 2008 Oct;7(10):4326-35. doi: 10.1021/pr800260a. Epub 2008 Aug 28. J Proteome Res. 2008. PMID: 18754630
-
Middle-Down and Top-Down Mass Spectrometric Analysis of Co-occurring Histone Modifications.Curr Protoc Protein Sci. 2014 Aug 1;77:23.7.1-23.7.28. doi: 10.1002/0471140864.ps2307s77. Curr Protoc Protein Sci. 2014. PMID: 25081742 Free PMC article. Review.
-
Characterization of histones and their post-translational modifications by mass spectrometry.Curr Opin Chem Biol. 2007 Feb;11(1):66-73. doi: 10.1016/j.cbpa.2006.11.022. Epub 2006 Dec 6. Curr Opin Chem Biol. 2007. PMID: 17157550 Review.
Cited by
-
Top-down proteomics in health and disease: challenges and opportunities.Proteomics. 2014 May;14(10):1195-210. doi: 10.1002/pmic.201300432. Proteomics. 2014. PMID: 24723472 Free PMC article. Review.
-
Mass spectrometry-based proteomics for the analysis of chromatin structure and dynamics.Int J Mol Sci. 2013 Mar 6;14(3):5402-31. doi: 10.3390/ijms14035402. Int J Mol Sci. 2013. PMID: 23466885 Free PMC article.
-
Quantitative Proteomics Reveals Fundamental Regulatory Differences in Oncogenic HRAS and Isocitrate Dehydrogenase (IDH1) Driven Astrocytoma.Mol Cell Proteomics. 2017 Jan;16(1):39-56. doi: 10.1074/mcp.M116.063883. Epub 2016 Nov 10. Mol Cell Proteomics. 2017. PMID: 27834733 Free PMC article.
-
Peptide Sequence Analysis by Electron Transfer Dissociation Mass Spectrometry: A Web-Based Tutorial.J Am Soc Mass Spectrom. 2015 Jul;26(7):1256-8. doi: 10.1007/s13361-015-1105-2. Epub 2015 Mar 28. J Am Soc Mass Spectrom. 2015. PMID: 25821049 Free PMC article.
-
Revealing histone variant induced changes via quantitative proteomics.Crit Rev Biochem Mol Biol. 2011 Aug;46(4):284-94. doi: 10.3109/10409238.2011.577052. Crit Rev Biochem Mol Biol. 2011. PMID: 21526979 Free PMC article. Review.
References
-
- Burlingame A. L. (ed) (2005) Methods in Enzymology, Vol. 402, Elsevier Academic Press, San Diego, CA
-
- Gstaiger M., Aebersold R. (2009) Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat. Rev. Genet 10, 617–627 - PubMed
-
- Cravatt B. F., Simon G. M., Yates J. R., 3rd (2007) The biological impact of mass-spectrometry-based proteomics. Nature 450, 991–1000 - PubMed
-
- Yates J. R., Ruse C. I., Nakorchevsky A. (2009) Proteomics by mass spectrometry: approaches, advances, and applications. Annu. Rev. Biomed. Eng 11, 49–79 - PubMed
-
- Zubarev R. A., Kelleher N. L., McLafferty F. W. (1998) Electron capture dissociation of multiply charged protein cations. A nonergodic process. J. Am. Chem. Soc 120, 3265–3266
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

