Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor
- PMID: 20124551
- PMCID: PMC2819218
- DOI: 10.1126/scisignal.2000568
Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor
Abstract
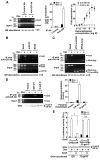
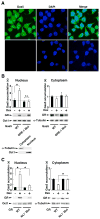
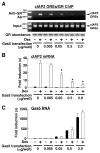
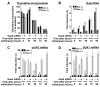
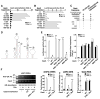
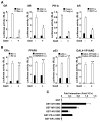
The availability of nutrients influences cellular growth and survival by affecting gene transcription. Glucocorticoids also influence gene transcription and have diverse activities on cell growth, energy expenditure, and survival. We found that the growth arrest-specific 5 (Gas5) noncoding RNA, which is abundant in cells whose growth has been arrested because of lack of nutrients or growth factors, sensitized cells to apoptosis by suppressing glucocorticoid-mediated induction of several responsive genes, including the one encoding cellular inhibitor of apoptosis 2. Gas5 bound to the DNA-binding domain of the glucocorticoid receptor (GR) by acting as a decoy glucocorticoid response element (GRE), thus competing with DNA GREs for binding to the GR. We conclude that Gas5 is a "riborepressor" of the GR, influencing cell survival and metabolic activities during starvation by modulating the transcriptional activity of the GR.
Figures


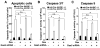

Similar articles
-
The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells.PLoS One. 2013;8(1):e55684. doi: 10.1371/journal.pone.0055684. Epub 2013 Jan 30. PLoS One. 2013. PMID: 23383264 Free PMC article.
-
Differential Expression of Glucocorticoid Receptor Noncoding RNA Repressor Gas5 in Autoimmune and Inflammatory Diseases.Horm Metab Res. 2016 Aug;48(8):550-7. doi: 10.1055/s-0042-106898. Epub 2016 May 23. Horm Metab Res. 2016. PMID: 27214311
-
Long noncoding RNA GAS5: a novel marker involved in glucocorticoid response.Curr Mol Med. 2015;15(1):94-9. doi: 10.2174/1566524015666150114122354. Curr Mol Med. 2015. PMID: 25601472
-
Negative glucocorticoid receptor response elements and their role in glucocorticoid action.Curr Pharm Des. 2004;10(23):2807-16. doi: 10.2174/1381612043383601. Curr Pharm Des. 2004. PMID: 15379669 Review.
-
Epigenetic Mechanisms of the Glucocorticoid Receptor.Trends Endocrinol Metab. 2019 Nov;30(11):807-818. doi: 10.1016/j.tem.2019.07.003. Trends Endocrinol Metab. 2019. PMID: 31699238 Review.
Cited by
-
Long noncoding RNA involvement in cancer.BMB Rep. 2012 Nov;45(11):604-11. doi: 10.5483/bmbrep.2012.45.11.227. BMB Rep. 2012. PMID: 23186998 Free PMC article. Review.
-
The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression.Genome Res. 2012 Sep;22(9):1775-89. doi: 10.1101/gr.132159.111. Genome Res. 2012. PMID: 22955988 Free PMC article.
-
LINC01116-dependent upregulation of RNA polymerase I transcription drives oncogenic phenotypes in lung adenocarcinoma.J Transl Med. 2024 Oct 5;22(1):904. doi: 10.1186/s12967-024-05715-5. J Transl Med. 2024. PMID: 39369230 Free PMC article.
-
Association of genetic variants in lncRNA GAS5/miR-21/mTOR axis with risk and prognosis of coronary artery disease among a Chinese population.J Clin Lab Anal. 2020 Oct;34(10):e23430. doi: 10.1002/jcla.23430. Epub 2020 Jun 17. J Clin Lab Anal. 2020. PMID: 32557866 Free PMC article.
-
Long noncoding RNAs in biology and hematopoiesis.Blood. 2013 Jun 13;121(24):4842-6. doi: 10.1182/blood-2013-03-456111. Epub 2013 May 3. Blood. 2013. PMID: 23645840 Free PMC article. Review.
References
-
- Lopez-Maury L, Marguerat S, Bahler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet. 2008;9:583–593. - PubMed
-
- Sellick CA, Reece RJ. Eukaryotic transcription factors as direct nutrient sensors. Trends Biochem Sci. 2005;30:405–412. - PubMed
-
- Kino T, Chrousos GP. Glucocorticoid Effect on Gene Expression. In: Steckler T, Kalin NH, Reul JMHM, editors. Handbook on Stress and the Brain Part 1. Elsevier BV; Amsterdam: 2004. pp. 295–312.
-
- Chrousos GP. Glucocorticoid therapy. In: Felig P, Frohman LA, editors. Endocrinology & Metablism. McGraw-Hill; New York: 2001. pp. 609–632.
-
- Chrousos GP, Kino T. Intracellular glucocorticoid signaling: a formerly simple system turns stochastic. Sci STKE 2005. 2005:pe48. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

