Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae
- PMID: 20118937
- PMCID: PMC2840703
- DOI: 10.1038/nsmb.1734
Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae
Abstract
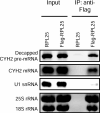
Nonsense-mediated decay (NMD) degrades mRNA containing premature translation termination codons. In yeast, NMD substrates are decapped and digested exonucleolytically from the 5' end. Despite the requirement for translation in recognition, degradation of nonsense-containing mRNA is considered to occur in ribosome-free cytoplasmic P bodies. We show decapped nonsense-containing mRNA associate with polyribosomes, indicating that recognition and degradation are tightly coupled and that polyribosomes are major sites for degradation of aberrant mRNAs.
Figures
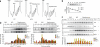
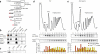
Similar articles
-
Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae.RNA. 1997 Mar;3(3):234-44. RNA. 1997. PMID: 9056761 Free PMC article.
-
ATP hydrolysis by UPF1 is required for efficient translation termination at premature stop codons.Nat Commun. 2016 Dec 23;7:14021. doi: 10.1038/ncomms14021. Nat Commun. 2016. PMID: 28008922 Free PMC article.
-
Targeting of aberrant mRNAs to cytoplasmic processing bodies.Cell. 2006 Jun 16;125(6):1095-109. doi: 10.1016/j.cell.2006.04.037. Cell. 2006. PMID: 16777600 Free PMC article.
-
Cutting the nonsense: the degradation of PTC-containing mRNAs.Biochem Soc Trans. 2010 Dec;38(6):1615-20. doi: 10.1042/BST0381615. Biochem Soc Trans. 2010. PMID: 21118136 Review.
-
Nonsense-mediated mRNA decay in Saccharomyces cerevisiae.Gene. 2001 Aug 22;274(1-2):15-25. doi: 10.1016/s0378-1119(01)00552-2. Gene. 2001. PMID: 11674994 Review.
Cited by
-
A eukaryotic translation initiation factor 4E-binding protein promotes mRNA decapping and is required for PUF repression.Mol Cell Biol. 2012 Oct;32(20):4181-94. doi: 10.1128/MCB.00483-12. Epub 2012 Aug 13. Mol Cell Biol. 2012. PMID: 22890846 Free PMC article.
-
A quantitative assay for measuring mRNA decapping by splinted ligation reverse transcription polymerase chain reaction: qSL-RT-PCR.RNA. 2011 Mar;17(3):535-43. doi: 10.1261/rna.2436411. Epub 2011 Jan 10. RNA. 2011. PMID: 21220548 Free PMC article.
-
Loss of nonsense mediated decay suppresses mutations in Saccharomyces cerevisiae TRA1.BMC Genet. 2012 Mar 22;13:19. doi: 10.1186/1471-2156-13-19. BMC Genet. 2012. PMID: 22439631 Free PMC article.
-
RNA degradomes reveal substrates and importance for dark and nitrogen stress responses of Arabidopsis XRN4.Nucleic Acids Res. 2019 Sep 26;47(17):9216-9230. doi: 10.1093/nar/gkz712. Nucleic Acids Res. 2019. PMID: 31428786 Free PMC article.
-
Eukaryotic mRNA decapping factors: molecular mechanisms and activity.FEBS J. 2023 Nov;290(21):5057-5085. doi: 10.1111/febs.16626. Epub 2022 Sep 30. FEBS J. 2023. PMID: 36098474 Free PMC article. Review.
References
-
- Isken O, Maquat LE. Genes Dev. 2007;21:1833–1856. - PubMed
-
- Behm-Ansmant I, et al. FEBS Lett. 2007;581:2845–2853. - PubMed
-
- Amrani N, Sachs MS, Jacobson A. Nat. Rev. Mol. Cell Biol. 2006;7:415–425. - PubMed
-
- Baker KE, Parker R. Curr. Opin. Cell Biol. 2004;16:293–299. - PubMed
-
- Muhlrad D, Parker R. Nature. 1994;370:578–581. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

