Biphasic recruitment of transcriptional repressors to the murine cytomegalovirus major immediate-early promoter during the course of infection in vivo
- PMID: 20106920
- PMCID: PMC2838119
- DOI: 10.1128/JVI.02380-09
Biphasic recruitment of transcriptional repressors to the murine cytomegalovirus major immediate-early promoter during the course of infection in vivo
Abstract
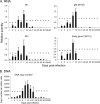
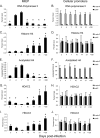
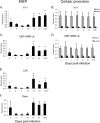
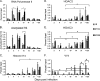
Our previous studies showed that establishment of murine cytomegalovirus (MCMV) latency in vivo is associated with repression of immediate-early gene expression, deacetylation of histones bound to the major immediate-early promoter (MIEP), changes in patterns of methylation of histones, and recruitment of cellular repressors of transcription to the MIEP. Here, we have quantitatively analyzed the kinetics of changes in viral RNA expression, DNA copy number, and recruitment of repressors and activators of transcription to viral promoters during the course of infection. Our results show that changes in viral gene expression correlate with changes in recruitment of RNA polymerase and acetylated histones to viral promoters. Binding of the transcriptional repressors histone deacetylase type 2 (HDAC2), HDAC3, YY1, CBF-1/RBP-Jk, Daxx, and CIR to the MIEP and HDACs to other promoters showed a biphasic pattern: some binding was detectable prior to activation of viral gene expression, then decreased with the onset of transcription and increased again as repression of viral gene expression occurred. Potential binding sites for CBF-1/RBP-Jk and YY1 in the MIEP and for YY1 in the M100 promoter (M100P) were identified by in silico analysis. While recruitment of HDACs was not promoter specific, binding of CBF-1/RBP-Jk and YY1 was restricted to promoters with their cognate sites. Our results suggest that sequences within viral promoters may contribute to establishment of latency through recruitment of transcriptional repressors to these genes. The observation that repressors are bound to the MIEP and other promoters immediately upon infection suggests that latency may be established in some cells very early in infection.
Figures
Similar articles
-
Establishment of murine cytomegalovirus latency in vivo is associated with changes in histone modifications and recruitment of transcriptional repressors to the major immediate-early promoter.J Virol. 2008 Nov;82(21):10922-31. doi: 10.1128/JVI.00865-08. Epub 2008 Aug 27. J Virol. 2008. PMID: 18753203 Free PMC article.
-
Control of cytomegalovirus lytic gene expression by histone acetylation.EMBO J. 2002 Mar 1;21(5):1112-20. doi: 10.1093/emboj/21.5.1112. EMBO J. 2002. PMID: 11867539 Free PMC article.
-
Chromatin-mediated regulation of cytomegalovirus gene expression.Virus Res. 2011 May;157(2):134-43. doi: 10.1016/j.virusres.2010.09.019. Epub 2010 Sep 25. Virus Res. 2011. PMID: 20875471 Free PMC article. Review.
-
Mouse cytomegalovirus immediate-early protein 1 binds with host cell repressors to relieve suppressive effects on viral transcription and replication during lytic infection.J Virol. 2003 Jan;77(2):1357-67. doi: 10.1128/jvi.77.2.1357-1367.2003. J Virol. 2003. PMID: 12502852 Free PMC article.
-
Chromatin structure regulates human cytomegalovirus gene expression during latency, reactivation and lytic infection.Biochim Biophys Acta. 2010 Mar-Apr;1799(3-4):286-95. doi: 10.1016/j.bbagrm.2009.08.001. Epub 2009 Aug 12. Biochim Biophys Acta. 2010. PMID: 19682613 Review.
Cited by
-
Bioactive Molecules Released From Cells Infected with the Human Cytomegalovirus.Front Microbiol. 2016 May 13;7:715. doi: 10.3389/fmicb.2016.00715. eCollection 2016. Front Microbiol. 2016. PMID: 27242736 Free PMC article. Review.
-
New Insights Into the Molecular Mechanisms and Immune Control of Cytomegalovirus Reactivation.Transplantation. 2020 May;104(5):e118-e124. doi: 10.1097/TP.0000000000003138. Transplantation. 2020. PMID: 31996662 Free PMC article. Review.
-
Transplant-induced reactivation of murine cytomegalovirus immediate early gene expression is associated with recruitment of NF-κB and AP-1 to the major immediate early promoter.J Gen Virol. 2016 Apr;97(4):941-954. doi: 10.1099/jgv.0.000407. Epub 2016 Jan 20. J Gen Virol. 2016. PMID: 26795571 Free PMC article.
-
Transient CD8-memory contraction: a potential contributor to latent cytomegalovirus reactivation.J Leukoc Biol. 2012 Nov;92(5):933-7. doi: 10.1189/jlb.1211635. Epub 2012 Jun 22. J Leukoc Biol. 2012. PMID: 22730545 Free PMC article.
-
Real-time transcriptional profiling of cellular and viral gene expression during lytic cytomegalovirus infection.PLoS Pathog. 2012 Sep;8(9):e1002908. doi: 10.1371/journal.ppat.1002908. Epub 2012 Sep 6. PLoS Pathog. 2012. PMID: 22969428 Free PMC article.
References
-
- Ahn, J. H., E. J. Brignole III, and G. S. Hayward. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol. 18:4899-4913. - PMC - PubMed
-
- Austen, M., B. Luscher, and J. M. Luscher-Firzlaff. 1997. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J. Biol. Chem. 272:1709-1717. - PubMed
-
- Bain, M., M. Reeves, and J. Sinclair. 2006. Regulation of human cytomegalovirus gene expression by chromatin remodeling, p. 167-183. In M. Reddehase (ed.), Cytomegaloviruses: molecular biology and immunology. Caister Academic Press, Norfolk, UK.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

