Hermansky-Pudlak protein complexes, AP-3 and BLOC-1, differentially regulate presynaptic composition in the striatum and hippocampus
- PMID: 20089890
- PMCID: PMC2824551
- DOI: 10.1523/JNEUROSCI.3400-09.2010
Hermansky-Pudlak protein complexes, AP-3 and BLOC-1, differentially regulate presynaptic composition in the striatum and hippocampus
Abstract
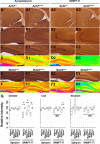
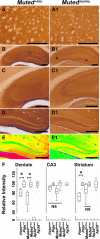
Endosomal sorting mechanisms mediated by AP-3 and BLOC-1 are perturbed in Hermansky-Pudlak Syndrome, a human genetic condition characterized by albinism and prolonged bleeding (OMIM #203300). Additionally, mouse models defective in either one of these complexes possess defective synaptic vesicle biogenesis (Newell-Litwa et al., 2009). These synaptic vesicle phenotypes were presumed uniform throughout the brain. However, here we report that AP-3 and BLOC-1 differentially regulate the composition of presynaptic terminals in the striatum and dentate gyrus of the hippocampus. Quantitative immunoelectron microscopy demonstrated that the majority of AP-3 immunoreactivity in both wild-type striatum and hippocampus localizes to presynaptic axonal compartments, where it regulates synaptic vesicle size. In the striatum, loss of AP-3 (Ap3d(mh/mh)) resulted in decreased synaptic vesicle size. In contrast, loss of AP-3 in the dentate gyrus increased synaptic vesicle size, thus suggesting anatomically specific AP-3-regulatory mechanisms. Loss-of-function alleles of BLOC-1, Pldn(pa/pa), and Muted(mu/mu) revealed that this complex acts as a brain-region-specific regulator of AP-3. In fact, BLOC-1 deficiencies selectively reduced AP-3 and AP-3 cargo immunoreactivity in presynaptic compartments within the dentate gyrus both at the light and/or electron microscopy level. However, the striatum did not exhibit these BLOC-1-null phenotypes. Our results demonstrate that distinct brain regions differentially regulate AP-3-dependent synaptic vesicle biogenesis. We propose that anatomically restricted mechanisms within the brain diversify the biogenesis and composition of synaptic vesicles.
Figures




Similar articles
-
Roles of BLOC-1 and adaptor protein-3 complexes in cargo sorting to synaptic vesicles.Mol Biol Cell. 2009 Mar;20(5):1441-53. doi: 10.1091/mbc.e08-05-0456. Epub 2009 Jan 14. Mol Biol Cell. 2009. PMID: 19144828 Free PMC article.
-
The zinc transporter ZnT3 interacts with AP-3 and it is preferentially targeted to a distinct synaptic vesicle subpopulation.Mol Biol Cell. 2004 Feb;15(2):575-87. doi: 10.1091/mbc.e03-06-0401. Epub 2003 Dec 2. Mol Biol Cell. 2004. PMID: 14657250 Free PMC article.
-
The neural cell adhesion molecule promotes maturation of the presynaptic endocytotic machinery by switching synaptic vesicle recycling from adaptor protein 3 (AP-3)- to AP-2-dependent mechanisms.J Neurosci. 2013 Oct 16;33(42):16828-45. doi: 10.1523/JNEUROSCI.2192-13.2013. J Neurosci. 2013. PMID: 24133283 Free PMC article.
-
Direct interaction of Dysbindin with the AP-3 complex via its mu subunit.Neurochem Int. 2009 Jun;54(7):431-8. doi: 10.1016/j.neuint.2009.01.014. Epub 2009 Jan 31. Neurochem Int. 2009. PMID: 19428785
-
What is the function of neuronal AP-3?Biol Cell. 2007 Jul;99(7):349-61. doi: 10.1042/BC20070029. Biol Cell. 2007. PMID: 17567262 Review.
Cited by
-
Hermansky-Pudlak syndrome: Mutation update.Hum Mutat. 2020 Mar;41(3):543-580. doi: 10.1002/humu.23968. Epub 2020 Jan 23. Hum Mutat. 2020. PMID: 31898847 Free PMC article.
-
Dysbindin-1C is required for the survival of hilar mossy cells and the maturation of adult newborn neurons in dentate gyrus.J Biol Chem. 2014 Oct 17;289(42):29060-72. doi: 10.1074/jbc.M114.590927. Epub 2014 Aug 25. J Biol Chem. 2014. PMID: 25157109 Free PMC article.
-
The WASH complex, an endosomal Arp2/3 activator, interacts with the Hermansky-Pudlak syndrome complex BLOC-1 and its cargo phosphatidylinositol-4-kinase type IIα.Mol Biol Cell. 2013 Jul;24(14):2269-84. doi: 10.1091/mbc.E13-02-0088. Epub 2013 May 15. Mol Biol Cell. 2013. PMID: 23676666 Free PMC article.
-
Overlapping Machinery in Lysosome-Related Organelle Trafficking: A Lesson from Rare Multisystem Disorders.Cells. 2022 Nov 21;11(22):3702. doi: 10.3390/cells11223702. Cells. 2022. PMID: 36429129 Free PMC article. Review.
-
VGLUT2 Trafficking Is Differentially Regulated by Adaptor Proteins AP-1 and AP-3.Front Cell Neurosci. 2017 Oct 26;11:324. doi: 10.3389/fncel.2017.00324. eCollection 2017. Front Cell Neurosci. 2017. PMID: 29123471 Free PMC article.
References
-
- Baude A, Nusser Z, Molnár E, McIlhinney RA, Somogyi P. High-resolution immunogold localization of AMPA type glutamate receptor subunits at synaptic and non-synaptic sites in rat hippocampus. Neuroscience. 1995;69:1031–1055. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
