A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics
- PMID: 20087442
- PMCID: PMC2808052
- DOI: 10.7150/ijbs.6.51
A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics
Abstract
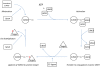
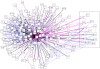
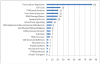
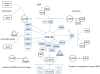
Promyelocytic Leukaemia Protein nuclear bodies (PML-NBs) are dynamic nuclear protein aggregates. To gain insight in PML-NB function, reductionist and high throughput techniques have been employed to identify PML-NB proteins. Here we present a manually curated network of the PML-NB interactome based on extensive literature review including database information. By compiling 'the PML-ome', we highlighted the presence of interactors in the Small Ubiquitin Like Modifier (SUMO) conjugation pathway. Additionally, we show an enrichment of SUMOylatable proteins in the PML-NBs through an in-house prediction algorithm. Therefore, based on the PML network, we hypothesize that PML-NBs may function as a nuclear SUMOylation hotspot.
Keywords: Cytoscape; PML-NB; SUMOylation; network; protein-protein interaction.
Conflict of interest statement
Conflict of Interest: The authors have declared that no conflict of interest exists.
Figures
Similar articles
-
Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins.J Cell Biol. 2014 Mar 17;204(6):931-45. doi: 10.1083/jcb.201305148. J Cell Biol. 2014. PMID: 24637324 Free PMC article.
-
SUMOylation regulates the number and size of promyelocytic leukemia-nuclear bodies (PML-NBs) and arsenic perturbs SUMO dynamics on PML by insolubilizing PML in THP-1 cells.Arch Toxicol. 2022 Feb;96(2):545-558. doi: 10.1007/s00204-021-03195-w. Epub 2022 Jan 10. Arch Toxicol. 2022. PMID: 35001170
-
The SUMO protease SENP6 is a direct regulator of PML nuclear bodies.Mol Biol Cell. 2011 Jan 1;22(1):78-90. doi: 10.1091/mbc.E10-06-0504. Epub 2010 Dec 9. Mol Biol Cell. 2011. PMID: 21148299 Free PMC article.
-
Unravelling the molecular interplay: SUMOylation, PML nuclear bodies and vascular cell activity in health and disease.Cell Signal. 2024 Jul;119:111156. doi: 10.1016/j.cellsig.2024.111156. Epub 2024 Apr 2. Cell Signal. 2024. PMID: 38574938 Review.
-
PML nuclear bodies and their spatial relationships in the mammalian cell nucleus.Front Biosci (Landmark Ed). 2009 Jan 1;14(3):1182-96. doi: 10.2741/3302. Front Biosci (Landmark Ed). 2009. PMID: 19273124 Review.
Cited by
-
On the Prevalence and Roles of Proteins Undergoing Liquid-Liquid Phase Separation in the Biogenesis of PML-Bodies.Biomolecules. 2023 Dec 18;13(12):1805. doi: 10.3390/biom13121805. Biomolecules. 2023. PMID: 38136675 Free PMC article.
-
Requirement of PML SUMO interacting motif for RNF4- or arsenic trioxide-induced degradation of nuclear PML isoforms.PLoS One. 2012;7(9):e44949. doi: 10.1371/journal.pone.0044949. Epub 2012 Sep 18. PLoS One. 2012. PMID: 23028697 Free PMC article.
-
Discrete foci containing RNase A are found in nucleoli of HeLa cells after aging in culture.Eur J Histochem. 2011 Apr 13;55(2):e15. doi: 10.4081/ejh.2011.e15. Eur J Histochem. 2011. PMID: 22193295 Free PMC article.
-
PML body meets telomere: the beginning of an ALTernate ending?Nucleus. 2012 May-Jun;3(3):263-75. doi: 10.4161/nucl.20326. Epub 2012 May 1. Nucleus. 2012. PMID: 22572954 Free PMC article. Review.
-
Dengue Non-structural Protein 5 Polymerase Complexes With Promyelocytic Leukemia Protein (PML) Isoforms III and IV to Disrupt PML-Nuclear Bodies in Infected Cells.Front Cell Infect Microbiol. 2019 Aug 13;9:284. doi: 10.3389/fcimb.2019.00284. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31456950 Free PMC article.
References
-
- de THE H, RIVIERE M, BERNHARD W. [Examination by electron microscope of the VX2 tumor of the domestic rabbit derived from the Shope papilloma.] Bull Assoc Fr Etud Cancer. 1960 Oct;47:570–84. - PubMed
-
- HINGLAIS-GUILLAUD N, MORICARD R, BERNHARD W. [Ultrastructure of invasive pavement-cell cancers of the uterine cervix in women.] Bull Assoc Fr Etud Cancer. 1961 Apr;48:283–316. - PubMed
-
- Jensen K, Shiels C, Freemont PS. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 2001 Oct 29;20(49):7223–33. - PubMed
-
- Borden KL. RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem Cell Biol. 1998;76(2-3):351–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

