Origin of irreversibility of cell cycle start in budding yeast
- PMID: 20087409
- PMCID: PMC2797597
- DOI: 10.1371/journal.pbio.1000284
Origin of irreversibility of cell cycle start in budding yeast
Erratum in
- PLoS Biol. 2011 Jul;9(7). doi:10.1371/annotation/90916531-1b34-4eb5-92a9-a7f8c5d72318
Abstract
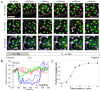
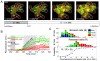
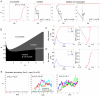
Budding yeast cells irreversibly commit to a new division cycle at a regulatory transition called Start. This essential decision-making step involves the activation of the SBF/MBF transcription factors. SBF/MBF promote expression of the G1 cyclins encoded by CLN1 and CLN2. Cln1,2 can activate their own expression by inactivating the Whi5 repressor of SBF/MBF. The resulting transcriptional positive feedback provides an appealing, but as yet unproven, candidate for generating irreversibility of Start. Here, we investigate the logic of the Start regulatory module by quantitative single-cell time-lapse microscopy, using strains in which expression of key regulators is efficiently controlled by changes of inducers in a microfluidic chamber. We show that Start activation is ultrasensitive to G1 cyclin. In the absence of CLN1,2-dependent positive feedback, we observe that Start transit is reversible, due to reactivation of the Whi5 transcriptional repressor. Introduction of the positive feedback loop makes Whi5 inactivation and Start activation irreversible, which therefore guarantees unidirectional entry into S phase. A simple mathematical model to describe G1 cyclin turn on at Start, entirely constrained by empirically measured parameters, shows that the experimentally measured ultrasensitivity and transcriptional positive feedback are necessary and sufficient dynamical characteristics to make the Start transition a bistable and irreversible switch. Our study thus demonstrates that Start irreversibility is a property that arises from the architecture of the system (Whi5/SBF/Cln2 loop), rather than the consequence of the regulation of a single component (e.g., irreversible protein degradation).
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
G1/S Transcription Factor Copy Number Is a Growth-Dependent Determinant of Cell Cycle Commitment in Yeast.Cell Syst. 2018 May 23;6(5):539-554.e11. doi: 10.1016/j.cels.2018.04.012. Cell Syst. 2018. PMID: 29792825
-
Modeling the START transition in the budding yeast cell cycle.PLoS Comput Biol. 2024 Aug 2;20(8):e1012048. doi: 10.1371/journal.pcbi.1012048. eCollection 2024 Aug. PLoS Comput Biol. 2024. PMID: 39093881 Free PMC article.
-
Dual regulation by pairs of cyclin-dependent protein kinases and histone deacetylases controls G1 transcription in budding yeast.PLoS Biol. 2009 Sep;7(9):e1000188. doi: 10.1371/journal.pbio.1000188. Epub 2009 Sep 8. PLoS Biol. 2009. PMID: 19823668 Free PMC article.
-
Is START a switch?Ciba Found Symp. 1992;170:20-5; discussion 25-9. doi: 10.1002/9780470514320.ch3. Ciba Found Symp. 1992. PMID: 1483346 Review.
-
MAPK cell-cycle regulation in Saccharomyces cerevisiae and Candida albicans.Future Microbiol. 2010 Jul;5(7):1125-41. doi: 10.2217/fmb.10.72. Future Microbiol. 2010. PMID: 20632810 Review.
Cited by
-
Role of a Candida albicans Nrm1/Whi5 homologue in cell cycle gene expression and DNA replication stress response.Mol Microbiol. 2012 May;84(4):778-94. doi: 10.1111/j.1365-2958.2012.08056.x. Epub 2012 Apr 16. Mol Microbiol. 2012. PMID: 22463761 Free PMC article.
-
Oscillatory dynamics of cell cycle proteins in single yeast cells analyzed by imaging cytometry.PLoS One. 2011;6(10):e26272. doi: 10.1371/journal.pone.0026272. Epub 2011 Oct 26. PLoS One. 2011. PMID: 22046265 Free PMC article.
-
Whi7 is an unstable cell-cycle repressor of the Start transcriptional program.Nat Commun. 2017 Aug 24;8(1):329. doi: 10.1038/s41467-017-00374-1. Nat Commun. 2017. PMID: 28839131 Free PMC article.
-
Fdo1, Fkh1, Fkh2, and the Swi6-Mbp1 MBF complex regulate Mcd1 levels to impact eco1 rad61 cell growth in Saccharomyces cerevisiae.Genetics. 2024 Oct 7;228(2):iyae128. doi: 10.1093/genetics/iyae128. Genetics. 2024. PMID: 39110836 Free PMC article.
-
Cell cycle commitment in budding yeast emerges from the cooperation of multiple bistable switches.Open Biol. 2011 Nov;1(3):110009. doi: 10.1098/rsob.110009. Open Biol. 2011. PMID: 22645649 Free PMC article.
References
-
- Hartwell L. H, Culotti J, Pringle J. R, Reid B. J. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. - PubMed
-
- Hereford L. M, Hartwell L. H. Sequential gene function in the initiation of Saccharomyces cerevisiae DNA synthesis. J Mol Biol. 1974;84:445–461. - PubMed
-
- Morgan D. O. The cell cycle: principles of control. London (United Kingdom): Oxford University Press; 2007. 297
-
- Costanzo M, Nishikawa J. L, Tang X, Millman J. S, Schub O, et al. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117:899–913. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

