NUDT16 and ITPA play a dual protective role in maintaining chromosome stability and cell growth by eliminating dIDP/IDP and dITP/ITP from nucleotide pools in mammals
- PMID: 20081199
- PMCID: PMC2875033
- DOI: 10.1093/nar/gkp1250
NUDT16 and ITPA play a dual protective role in maintaining chromosome stability and cell growth by eliminating dIDP/IDP and dITP/ITP from nucleotide pools in mammals
Abstract
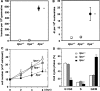
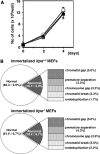
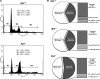
Mammalian inosine triphosphatase encoded by ITPA gene hydrolyzes ITP and dITP to monophosphates, avoiding their deleterious effects. Itpa(-) mice exhibited perinatal lethality, and significantly higher levels of inosine in cellular RNA and deoxyinosine in nuclear DNA were detected in Itpa(-) embryos than in wild-type embryos. Therefore, we examined the effects of ITPA deficiency on mouse embryonic fibroblasts (MEFs). Itpa(-) primary MEFs lacking ITP-hydrolyzing activity exhibited a prolonged doubling time, increased chromosome abnormalities and accumulation of single-strand breaks in nuclear DNA, compared with primary MEFs prepared from wild-type embryos. However, immortalized Itpa(-) MEFs had neither of these phenotypes and had a significantly higher ITP/IDP-hydrolyzing activity than Itpa(-) embryos or primary MEFs. Mammalian NUDT16 proteins exhibit strong dIDP/IDP-hydrolyzing activity and similarly low levels of Nudt16 mRNA and protein were detected in primary MEFs derived from both wild-type and Itpa(-) embryos. However, immortalized Itpa(-) MEFs expressed significantly higher levels of Nudt16 than the wild type. Moreover, introduction of silencing RNAs against Nudt16 into immortalized Itpa(-) MEFs reproduced ITPA-deficient phenotypes. We thus conclude that NUDT16 and ITPA play a dual protective role for eliminating dIDP/IDP and dITP/ITP from nucleotide pools in mammals.
Figures


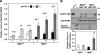
Similar articles
-
NUDT16 is a (deoxy)inosine diphosphatase, and its deficiency induces accumulation of single-strand breaks in nuclear DNA and growth arrest.Nucleic Acids Res. 2010 Aug;38(14):4834-43. doi: 10.1093/nar/gkq249. Epub 2010 Apr 12. Nucleic Acids Res. 2010. PMID: 20385596 Free PMC article.
-
ITPA protein, an enzyme that eliminates deaminated purine nucleoside triphosphates in cells.Mutat Res. 2010 Nov 28;703(1):43-50. doi: 10.1016/j.mrgentox.2010.06.009. Epub 2010 Jun 22. Mutat Res. 2010. PMID: 20601097 Review.
-
A comprehensive screening system for damaged nucleotide-binding proteins.Mutat Res. 2010 Nov 28;703(1):37-42. doi: 10.1016/j.mrgentox.2010.06.005. Epub 2010 Jun 11. Mutat Res. 2010. PMID: 20542141 Review.
-
Deoxyinosine triphosphate induces MLH1/PMS2- and p53-dependent cell growth arrest and DNA instability in mammalian cells.Sci Rep. 2016 Sep 13;6:32849. doi: 10.1038/srep32849. Sci Rep. 2016. PMID: 27618981 Free PMC article.
-
ITPA (inosine triphosphate pyrophosphatase): from surveillance of nucleotide pools to human disease and pharmacogenetics.Mutat Res. 2013 Oct-Dec;753(2):131-146. doi: 10.1016/j.mrrev.2013.08.001. Epub 2013 Aug 19. Mutat Res. 2013. PMID: 23969025 Free PMC article. Review.
Cited by
-
Genetic effect of an InDel in the promoter region of the NUDT15 and its effect on myoblast proliferation in chickens.BMC Genomics. 2022 Feb 16;23(1):138. doi: 10.1186/s12864-022-08362-6. BMC Genomics. 2022. PMID: 35168561 Free PMC article.
-
Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.Nat Struct Mol Biol. 2018 Jul;25(7):591-600. doi: 10.1038/s41594-018-0083-z. Epub 2018 Jul 2. Nat Struct Mol Biol. 2018. PMID: 29967538 Free PMC article.
-
Multiplatform tear proteomic profiling reveals novel non-invasive biomarkers for diabetic retinopathy.Eye (Lond). 2024 Jun;38(8):1509-1517. doi: 10.1038/s41433-024-02938-0. Epub 2024 Feb 9. Eye (Lond). 2024. PMID: 38336992
-
CAG RNAs induce DNA damage and apoptosis by silencing NUDT16 expression in polyglutamine degeneration.Proc Natl Acad Sci U S A. 2021 May 11;118(19):e2022940118. doi: 10.1073/pnas.2022940118. Proc Natl Acad Sci U S A. 2021. PMID: 33947817 Free PMC article.
-
Elevated Levels of DNA Strand Breaks Induced by a Base Analog in the Human Cell Line with the P32T ITPA Variant.J Nucleic Acids. 2010 Sep 26;2010:872180. doi: 10.4061/2010/872180. J Nucleic Acids. 2010. PMID: 20936128 Free PMC article.
References
-
- Nakabeppu Y, Tsuchimoto D, Furuichi M, Sakumi K. The defense mechanisms in mammalian cells against oxidative damage in nucleic acids and their involvement in the suppression of mutagenesis and cell death. Free Radic. Res. 2004;38:423–429. - PubMed
-
- Nakabeppu Y, Behmanesh M, Yamaguchi H, Yoshimura D, Sakumi K. In: Oxidative Damage to Nucleic Acids. Evans MD, Cooke MS, editors. TX/New York: Landes Bioscience/Springer, Austin; 2007. pp. 40–53.
-
- Nakabeppu Y. Molecular genetics and structural biology of human MutT homolog, MTH1. Mutat. Res. 2001;477:59–70. - PubMed
-
- Nakabeppu Y, Kajitani K, Sakamoto K, Yamaguchi H, Tsuchimoto D. MTH1, an oxidized purine nucleoside triphosphatase, prevents the cytotoxicity and neurotoxicity of oxidized purine nucleotides. DNA Rep. 2006;5:761–772. - PubMed
-
- Behmanesh M, Sakumi K, Tsuchimoto D, Torisu K, Ohnishi-Honda Y, Rancourt DE, Nakabeppu Y. Characterization of the structure and expression of mouse Itpa gene and its related sequences in the mouse genome. DNA Res. 2005;12:39–51. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

