KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming
- PMID: 20080955
- PMCID: PMC2807354
- DOI: 10.1101/gad.553410
KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming
Abstract
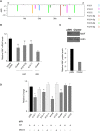
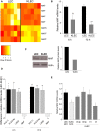
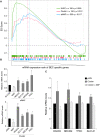
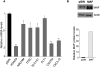
Kaposi sarcoma herpesvirus (KSHV) induces transcriptional reprogramming of endothelial cells. In particular, KSHV-infected lymphatic endothelial cells (LECs) show an up-regulation of genes associated with blood vessel endothelial cells (BECs). Consequently, KSHV-infected tumor cells in Kaposi sarcoma are poorly differentiated endothelial cells, expressing markers of both LECs and BECs. MicroRNAs (miRNAs) are short noncoding RNA molecules that act post-transcriptionally to negatively regulate gene expression. Here we validate expression of the KSHV-encoded miRNAs in Kaposi sarcoma lesions and demonstrate that these miRNAs contribute to viral-induced reprogramming by silencing the cellular transcription factor MAF (musculoaponeurotic fibrosarcoma oncogene homolog). MAF is expressed in LECs but not in BECs. We identify a novel role for MAF as a transcriptional repressor, preventing expression of BEC-specific genes, thereby maintaining the differentiation status of LECs. These findings demonstrate that viral miRNAs could influence the differentiation status of infected cells, and thereby contribute to KSHV-induced oncogenesis.
Figures



Similar articles
-
Lymphatic reprogramming by Kaposi sarcoma herpes virus promotes the oncogenic activity of the virus-encoded G-protein-coupled receptor.Cancer Res. 2012 Nov 15;72(22):5833-42. doi: 10.1158/0008-5472.CAN-12-1229. Epub 2012 Aug 31. Cancer Res. 2012. PMID: 22942256 Free PMC article.
-
Modified Cross-Linking, Ligation, and Sequencing of Hybrids (qCLASH) Identifies Kaposi's Sarcoma-Associated Herpesvirus MicroRNA Targets in Endothelial Cells.J Virol. 2018 Mar 28;92(8):e02138-17. doi: 10.1128/JVI.02138-17. Print 2018 Apr 15. J Virol. 2018. PMID: 29386283 Free PMC article.
-
Opposing regulation of PROX1 by interleukin-3 receptor and NOTCH directs differential host cell fate reprogramming by Kaposi sarcoma herpes virus.PLoS Pathog. 2012;8(6):e1002770. doi: 10.1371/journal.ppat.1002770. Epub 2012 Jun 14. PLoS Pathog. 2012. PMID: 22719258 Free PMC article.
-
The role of Kaposi sarcoma-associated herpesvirus in the pathogenesis of Kaposi sarcoma.J Pathol. 2015 Jan;235(2):368-80. doi: 10.1002/path.4441. J Pathol. 2015. PMID: 25212381 Review.
-
KSHV microRNAs: Tricks of the Devil.Trends Microbiol. 2017 Aug;25(8):648-661. doi: 10.1016/j.tim.2017.02.002. Epub 2017 Mar 2. Trends Microbiol. 2017. PMID: 28259385 Free PMC article. Review.
Cited by
-
Viral miRNA regulation of host gene expression.Semin Cell Dev Biol. 2023 Sep 15;146:2-19. doi: 10.1016/j.semcdb.2022.11.007. Epub 2022 Nov 30. Semin Cell Dev Biol. 2023. PMID: 36463091 Free PMC article. Review.
-
Hepatitis B Virus-Encoded MicroRNA (HBV-miR-3) Inhibits FIH-1 Expression to Promote Tumor Angiogenesis in HBV-Related Hepatocellular Carcinoma.J Hepatocell Carcinoma. 2023 Dec 27;10:2337-2353. doi: 10.2147/JHC.S436926. eCollection 2023. J Hepatocell Carcinoma. 2023. PMID: 38163053 Free PMC article.
-
The chromatin landscape of Kaposi's sarcoma-associated herpesvirus.Viruses. 2013 May 23;5(5):1346-73. doi: 10.3390/v5051346. Viruses. 2013. PMID: 23698402 Free PMC article. Review.
-
Dietary plant miRNAs as an augmented therapy: cross-kingdom gene regulation.RNA Biol. 2018;15(12):1433-1439. doi: 10.1080/15476286.2018.1551693. Epub 2018 Dec 10. RNA Biol. 2018. PMID: 30474479 Free PMC article. Review.
-
Functions of Kaposi's sarcoma-associated herpesvirus microRNAs.Biochim Biophys Acta. 2011 Nov-Dec;1809(11-12):623-30. doi: 10.1016/j.bbagrm.2011.05.003. Epub 2011 May 18. Biochim Biophys Acta. 2011. PMID: 21616184 Free PMC article. Review.
References
-
- Aziz A, Soucie E, Sarrazin S, Sieweke MH. MafB/c-Maf deficiency enables self-renewal of differentiated functional macrophages. Science. 2009;326:867–871. - PubMed
-
- Bixel MG, Adams RH. Master and commander: Continued expression of Prox1 prevents the dedifferentiation of lymphatic endothelial cells. Genes & Dev. 2008;22:3232–3235. - PubMed
-
- Boshoff C, Weiss R. AIDS-related malignancies. Nat Rev Cancer. 2002;2:373–382. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
