Dynein light chain 1 is required for autophagy, protein clearance, and cell death in Drosophila
- PMID: 20080745
- PMCID: PMC2818958
- DOI: 10.1073/pnas.0907967107
Dynein light chain 1 is required for autophagy, protein clearance, and cell death in Drosophila
Abstract
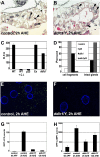
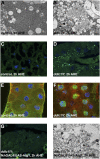
Autophagy is a catabolic pathway that is important for turnover of long-lived proteins and organelles, and has been implicated in cell survival, tumor progression, protection from infection, neurodegeneration, and cell death. Autophagy and caspases are required for type II autophagic cell death of Drosophila larval salivary glands during development, but the mechanisms that regulate these degradation pathways are not understood. We conducted a forward genetic screen for genes that are required for salivary gland cell death, and here we describe the identification of Drosophila dynein light chain 1 (ddlc1) as a gene that is required for type II cell death. Autophagy is attenuated in ddlc1 mutants, but caspases are active in these cells. ddlc1 mutant salivary glands develop large fibrillar protein inclusions that stain positive for amyloid-specific dyes and ubiquitin. Ectopic expression of Atg1 is sufficient to induce autophagy, clear protein inclusions, and rescue degradation of ddlc1 mutant salivary glands. Furthermore, ddlc1 mutant larvae have decreased motility, and mutations in ddlc1 enhance the impairment of motility that is observed in a Drosophila model of neurodegenerative disease. Significantly, this decrease in larval motility is associated with decreased clearance of protein with polyglutamine expansion, the accumulation of p62 in neurons and muscles, and fewer synaptic boutons. These results indicate that DDLC1 is required for protein clearance by autophagy that is associated with autophagic cell death and neurodegeneration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

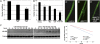

Similar articles
-
Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila.Cell. 2007 Dec 14;131(6):1137-48. doi: 10.1016/j.cell.2007.10.048. Cell. 2007. PMID: 18083103 Free PMC article.
-
Warts is required for PI3K-regulated growth arrest, autophagy, and autophagic cell death in Drosophila.Curr Biol. 2008 Oct 14;18(19):1466-75. doi: 10.1016/j.cub.2008.08.052. Epub 2008 Sep 25. Curr Biol. 2008. PMID: 18818081 Free PMC article.
-
Activation of autophagy during cell death requires the engulfment receptor Draper.Nature. 2010 Jun 24;465(7301):1093-6. doi: 10.1038/nature09127. Nature. 2010. PMID: 20577216 Free PMC article.
-
Autophagic programmed cell death in Drosophila.Cell Death Differ. 2003 Sep;10(9):940-5. doi: 10.1038/sj.cdd.4401280. Cell Death Differ. 2003. PMID: 12934068 Review.
-
Larval midgut destruction in Drosophila: not dependent on caspases but suppressed by the loss of autophagy.Autophagy. 2010 Jan;6(1):163-5. doi: 10.4161/auto.6.1.10601. Epub 2010 Jan 11. Autophagy. 2010. PMID: 20009534 Free PMC article. Review.
Cited by
-
The novel zinc finger protein dASCIZ regulates mitosis in Drosophila via an essential role in dynein light-chain expression.Genetics. 2014 Feb;196(2):443-53. doi: 10.1534/genetics.113.159541. Epub 2013 Dec 13. Genetics. 2014. PMID: 24336747 Free PMC article.
-
Inverse regulation of two classic Hippo pathway target genes in Drosophila by the dimerization hub protein Ctp.Sci Rep. 2016 Mar 14;6:22726. doi: 10.1038/srep22726. Sci Rep. 2016. PMID: 26972460 Free PMC article.
-
Rapid Genomic Evolution Drives the Diversification of Male Reproductive Genes in Dung Beetles.Genome Biol Evol. 2021 Aug 3;13(8):evab172. doi: 10.1093/gbe/evab172. Genome Biol Evol. 2021. PMID: 34426833 Free PMC article.
-
Valosin-containing protein is a key mediator between autophagic cell death and apoptosis in adult hippocampal neural stem cells following insulin withdrawal.Mol Brain. 2016 Mar 22;9:31. doi: 10.1186/s13041-016-0212-8. Mol Brain. 2016. PMID: 27000202 Free PMC article.
-
Dysregulated Dynein-Mediated Trafficking of Nephrin Causes INF2-related Podocytopathy.J Am Soc Nephrol. 2021 Feb;32(2):307-322. doi: 10.1681/ASN.2020081109. Epub 2020 Dec 22. J Am Soc Nephrol. 2021. PMID: 33443052 Free PMC article.
References
-
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. - PubMed
-
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. - PubMed
-
- Thumm M, et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

