Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis
- PMID: 20071528
- PMCID: PMC2821110
- DOI: 10.1523/JNEUROSCI.4988-09.2010
Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis
Abstract
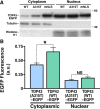
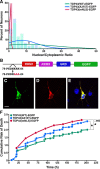
Mutations in the gene encoding TDP-43-the major protein component of neuronal aggregates characteristic of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) with ubiquitin-positive inclusion bodies-have been linked to familial forms of both disorders. Aggregates of TDP-43 in cortical and spinal motorneurons in ALS, or in neurons of the frontal and temporal cortices in FTLD, are closely linked to neuron loss and atrophy in these areas. However, the mechanism by which TDP-43 mutations lead to neurodegeneration is unclear. To investigate the pathogenic role of TDP-43 mutations, we established a model of TDP-43 proteinopathies by expressing fluorescently tagged wild-type and mutant TDP-43 in primary rat cortical neurons. Expression of mutant TDP-43 was toxic to neurons, and mutant-specific toxicity was associated with increased cytoplasmic mislocalization of TDP-43. Inclusion bodies were not necessary for the toxicity and did not affect the risk of cell death. Cellular survival was unaffected by the total amount of exogenous TDP-43 in the nucleus, but the amount of cytoplasmic TDP-43 was a strong and independent predictor of neuronal death. These results suggest that mutant TDP-43 is mislocalized to the cytoplasm, where it exhibits a toxic gain-of-function and induces cell death.
Figures

Similar articles
-
ALS/FTLD-linked TDP-43 regulates neurite morphology and cell survival in differentiated neurons.Exp Cell Res. 2013 Aug 1;319(13):1998-2005. doi: 10.1016/j.yexcr.2013.05.025. Epub 2013 Jun 4. Exp Cell Res. 2013. PMID: 23742895
-
[TDP-43 proteinopathies: ALS and frontotemporal dementias].Fortschr Neurol Psychiatr. 2009 Aug;77 Suppl 1:S25-7. doi: 10.1055/s-0028-1109602. Epub 2009 Aug 14. Fortschr Neurol Psychiatr. 2009. PMID: 19685386 German.
-
TARDBP pathogenic mutations increase cytoplasmic translocation of TDP-43 and cause reduction of endoplasmic reticulum Ca²⁺ signaling in motor neurons.Neurobiol Dis. 2015 Mar;75:64-77. doi: 10.1016/j.nbd.2014.12.010. Epub 2014 Dec 17. Neurobiol Dis. 2015. PMID: 25526708
-
[FTLD/ALS as TDP-43 proteinopathies].Rinsho Shinkeigaku. 2010 Nov;50(11):1022-4. doi: 10.5692/clinicalneurol.50.1022. Rinsho Shinkeigaku. 2010. PMID: 21921552 Review. Japanese.
-
[Clinical and pathological spectrum of TDP-43 associated ALS].Rinsho Shinkeigaku. 2010 Nov;50(11):940-2. doi: 10.5692/clinicalneurol.50.940. Rinsho Shinkeigaku. 2010. PMID: 21921519 Review. Japanese.
Cited by
-
Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration.Nat Rev Neurosci. 2011 Nov 30;13(1):38-50. doi: 10.1038/nrn3121. Nat Rev Neurosci. 2011. PMID: 22127299 Free PMC article. Review.
-
TARDBP mutation analysis in TDP-43 proteinopathies and deciphering the toxicity of mutant TDP-43.J Alzheimers Dis. 2013;33 Suppl 1(Suppl 1):S35-45. doi: 10.3233/JAD-2012-129036. J Alzheimers Dis. 2013. PMID: 22751173 Free PMC article. Review.
-
Mis-spliced transcripts generate de novo proteins in TDP-43-related ALS/FTD.Sci Transl Med. 2024 Feb 14;16(734):eadg7162. doi: 10.1126/scitranslmed.adg7162. Epub 2024 Feb 14. Sci Transl Med. 2024. PMID: 38277467 Free PMC article.
-
Respiratory pathology in the TDP-43 transgenic mouse model of amyotrophic lateral sclerosis.Front Physiol. 2024 Aug 27;15:1430875. doi: 10.3389/fphys.2024.1430875. eCollection 2024. Front Physiol. 2024. PMID: 39403566 Free PMC article.
-
Proteostasis in the Male and Female Germline: A New Outlook on the Maintenance of Reproductive Health.Front Cell Dev Biol. 2021 Apr 16;9:660626. doi: 10.3389/fcell.2021.660626. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33937261 Free PMC article. Review.
References
-
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, Oda T. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. - PubMed
-
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. - PubMed
-
- Ayala YM, Zago P, D'Ambrogio A, Xu YF, Petrucelli L, Buratti E, Baralle FE. Structural determinants of the cellular localization and shuttling of TDP-43. J Cell Sci. 2008;121:3778–3785. - PubMed
-
- Benajiba L, Le Ber I, Camuzat A, Lacoste M, Thomas-Anterion C, Couratier P, Legallic S, Salachas F, Hannequin D, Decousus M, Lacomblez L, Guedj E, Golfier V, Camu W, Dubois B, Campion D, Meininger V, Brice A French Clinical and Genetic Research Network on Frontotemporal Lobar Degeneration/Frontotemporal Lobar Degeneration with Motoneuron Disease. TARDBP mutations in motoneuron disease with frontotemporal lobar degeneration. Ann Neurol. 2009;65:470–473. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
