ETE: a python Environment for Tree Exploration
- PMID: 20070885
- PMCID: PMC2820433
- DOI: 10.1186/1471-2105-11-24
ETE: a python Environment for Tree Exploration
Abstract
Background: Many bioinformatics analyses, ranging from gene clustering to phylogenetics, produce hierarchical trees as their main result. These are used to represent the relationships among different biological entities, thus facilitating their analysis and interpretation. A number of standalone programs are available that focus on tree visualization or that perform specific analyses on them. However, such applications are rarely suitable for large-scale surveys, in which a higher level of automation is required. Currently, many genome-wide analyses rely on tree-like data representation and hence there is a growing need for scalable tools to handle tree structures at large scale.
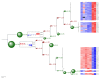
Results: Here we present the Environment for Tree Exploration (ETE), a python programming toolkit that assists in the automated manipulation, analysis and visualization of hierarchical trees. ETE libraries provide a broad set of tree handling options as well as specific methods to analyze phylogenetic and clustering trees. Among other features, ETE allows for the independent analysis of tree partitions, has support for the extended newick format, provides an integrated node annotation system and permits to link trees to external data such as multiple sequence alignments or numerical arrays. In addition, ETE implements a number of built-in analytical tools, including phylogeny-based orthology prediction and cluster validation techniques. Finally, ETE's programmable tree drawing engine can be used to automate the graphical rendering of trees with customized node-specific visualizations.
Conclusions: ETE provides a complete set of methods to manipulate tree data structures that extends current functionality in other bioinformatic toolkits of a more general purpose. ETE is free software and can be downloaded from http://ete.cgenomics.org.
Figures
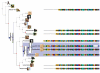
Similar articles
-
ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data.Mol Biol Evol. 2016 Jun;33(6):1635-8. doi: 10.1093/molbev/msw046. Epub 2016 Feb 26. Mol Biol Evol. 2016. PMID: 26921390 Free PMC article.
-
A Critical Review on the Use of Support Values in Tree Viewers and Bioinformatics Toolkits.Mol Biol Evol. 2017 Jun 1;34(6):1535-1542. doi: 10.1093/molbev/msx055. Mol Biol Evol. 2017. PMID: 28369572 Free PMC article.
-
PhyloCloud: an online platform for making sense of phylogenomic data.Nucleic Acids Res. 2022 Jul 5;50(W1):W577-W582. doi: 10.1093/nar/gkac324. Nucleic Acids Res. 2022. PMID: 35544233 Free PMC article.
-
Inferring trees.Methods Mol Biol. 2008;452:287-309. doi: 10.1007/978-1-60327-159-2_14. Methods Mol Biol. 2008. PMID: 18566770 Review.
-
PANTHER: Making genome-scale phylogenetics accessible to all.Protein Sci. 2022 Jan;31(1):8-22. doi: 10.1002/pro.4218. Epub 2021 Nov 25. Protein Sci. 2022. PMID: 34717010 Free PMC article. Review.
Cited by
-
Genetic Drift, Not Life History or RNAi, Determine Long-Term Evolution of Transposable Elements.Genome Biol Evol. 2016 Oct 5;8(9):2964-2978. doi: 10.1093/gbe/evw208. Genome Biol Evol. 2016. PMID: 27566762 Free PMC article.
-
Bio.Phylo: a unified toolkit for processing, analyzing and visualizing phylogenetic trees in Biopython.BMC Bioinformatics. 2012 Aug 21;13:209. doi: 10.1186/1471-2105-13-209. BMC Bioinformatics. 2012. PMID: 22909249 Free PMC article.
-
Characterization of integrated prophages within diverse species of clinical nontuberculous mycobacteria.Virol J. 2020 Aug 17;17(1):124. doi: 10.1186/s12985-020-01394-y. Virol J. 2020. PMID: 32807206 Free PMC article.
-
Phylogenetic Distribution of Secondary Metabolites in the Bacillus subtilis Species Complex.mSystems. 2021 Mar 9;6(2):e00057-21. doi: 10.1128/mSystems.00057-21. mSystems. 2021. PMID: 33688015 Free PMC article.
-
Molecular evolution and signatures of selective pressures on Bos, focusing on the Nelore breed (Bos indicus).PLoS One. 2022 Dec 22;17(12):e0279091. doi: 10.1371/journal.pone.0279091. eCollection 2022. PLoS One. 2022. PMID: 36548260 Free PMC article.
References
-
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12(4):357–358. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

