Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice
- PMID: 20062804
- PMCID: PMC2799519
- DOI: 10.1371/journal.pone.0008579
Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice
Abstract
Nuclear factor-erythroid 2 p45-related factor 2 (Nrf2) is a transcription factor which regulates the expression of many cytoprotective genes. In the present study, we found that the expression of Nrf2 was suppressed in prostate tumor of the Transgenic Adenocarcinoma of Mouse Prostate (TRAMP) mice. Similarly, the expression of Nrf2 and the induction of NQO1 were also substantially suppressed in tumorigenic TRAMP C1 cells but not in non-tumorigenic TRAMP C3 cells. Examination of the promoter region of the mouse Nrf2 gene identified a CpG island, which was methylated at specific CpG sites in prostate TRAMP tumor and in TRAMP C1 cells but not in normal prostate or TRAMP C3 cells, as shown by bisulfite genomic sequencing. Reporter assays indicated that methylation of these CpG sites dramatically inhibited the transcriptional activity of the Nrf2 promoter. Chromatin immunopreceipitation (ChIP) assays revealed increased binding of the methyl-CpG-binding protein 2 (MBD2) and trimethyl-histone H3 (Lys9) proteins to these CpG sites in the TRAMP C1 cells as compared to TRAMP C3 cells. In contrast, the binding of RNA Pol II and acetylated histone H3 to the Nrf2 promoter was decreased. Furthermore, treatment of TRAMP C1 cells with DNA methyltransferase (DNMT) inhibitor 5-aza-2'-deoxycytidine (5-aza) and histone deacetylase (HDAC) inhibitor trichostatin A (TSA) restored the expression of Nrf2 as well as the induction of NQO1 in TRAMP C1 cells. Taken together, these results indicate that the expression of Nrf2 is suppressed epigenetically by promoter methylation associated with MBD2 and histone modifications in the prostate tumor of TRAMP mice. Our present findings reveal a novel mechanism by which Nrf2 expression is suppressed in TRAMP prostate tumor, shed new light on the role of Nrf2 in carcinogenesis and provide potential new directions for the detection and prevention of prostate cancer.
Conflict of interest statement
Figures


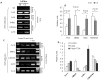
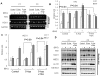
Similar articles
-
A γ-tocopherol-rich mixture of tocopherols maintains Nrf2 expression in prostate tumors of TRAMP mice via epigenetic inhibition of CpG methylation.J Nutr. 2012 May;142(5):818-23. doi: 10.3945/jn.111.153114. Epub 2012 Mar 28. J Nutr. 2012. PMID: 22457388 Free PMC article.
-
Epigenetic DNA methylation of antioxidative stress regulator NRF2 in human prostate cancer.Cancer Prev Res (Phila). 2014 Dec;7(12):1186-97. doi: 10.1158/1940-6207.CAPR-14-0127. Epub 2014 Sep 29. Cancer Prev Res (Phila). 2014. PMID: 25266896 Free PMC article.
-
The triterpenoid corosolic acid blocks transformation and epigenetically reactivates Nrf2 in TRAMP-C1 prostate cells.Mol Carcinog. 2018 Apr;57(4):512-521. doi: 10.1002/mc.22776. Epub 2018 Jan 11. Mol Carcinog. 2018. PMID: 29247555 Free PMC article.
-
NRF2 modulation in TRAMP mice: an in vivo model of prostate cancer.Mol Biol Rep. 2023 Jan;50(1):873-881. doi: 10.1007/s11033-022-08052-2. Epub 2022 Nov 6. Mol Biol Rep. 2023. PMID: 36335520 Review.
-
The Complex Genetic and Epigenetic Regulation of the Nrf2 Pathways: A Review.Antioxidants (Basel). 2023 Feb 3;12(2):366. doi: 10.3390/antiox12020366. Antioxidants (Basel). 2023. PMID: 36829925 Free PMC article. Review.
Cited by
-
Nrf2 Signaling Pathway: Focus on Oxidative Stress in Spinal Cord Injury.Mol Neurobiol. 2024 Aug 2. doi: 10.1007/s12035-024-04394-z. Online ahead of print. Mol Neurobiol. 2024. PMID: 39093381 Review.
-
Transcription Factor NRF2 in Shaping Myeloid Cell Differentiation and Function.Adv Exp Med Biol. 2024;1459:159-195. doi: 10.1007/978-3-031-62731-6_8. Adv Exp Med Biol. 2024. PMID: 39017844 Review.
-
The Role of NRF2 in Trinucleotide Repeat Expansion Disorders.Antioxidants (Basel). 2024 May 26;13(6):649. doi: 10.3390/antiox13060649. Antioxidants (Basel). 2024. PMID: 38929088 Free PMC article. Review.
-
Keap1-Nrf2 pathway: a key mechanism in the occurrence and development of cancer.Front Oncol. 2024 Apr 3;14:1381467. doi: 10.3389/fonc.2024.1381467. eCollection 2024. Front Oncol. 2024. PMID: 38634043 Free PMC article. Review.
-
The Contribution of Hippocampal All-Trans Retinoic Acid (ATRA) Deficiency to Alzheimer's Disease: A Narrative Overview of ATRA-Dependent Gene Expression in Post-Mortem Hippocampal Tissue.Antioxidants (Basel). 2023 Oct 27;12(11):1921. doi: 10.3390/antiox12111921. Antioxidants (Basel). 2023. PMID: 38001775 Free PMC article. Review.
References
-
- Yossepowitch O, Pinchuk I, Gur U, Neumann A, Lichtenberg D, et al. Advanced but not localized prostate cancer is associated with increased oxidative stress. J Urol. 2007;178:1238–1243; discussion 1243–1234. - PubMed
-
- Minelli A, Bellezza I, Conte C, Culig Z. Oxidative stress-related aging: A role for prostate cancer? Biochim Biophys Acta. 2009;1795:83–91. - PubMed
-
- Federico A, Morgillo F, Tuccillo C, Ciardiello F, Loguercio C. Chronic inflammation and oxidative stress in human carcinogenesis. Int J Cancer. 2007;121:2381–2386. - PubMed
-
- Yu S, Kong AN. Targeting carcinogen metabolism by dietary cancer preventive compounds. Curr Cancer Drug Targets. 2007;7:416–424. - PubMed
-
- Nakayama M, Gonzalgo ML, Yegnasubramanian S, Lin X, De Marzo AM, et al. GSTP1 CpG island hypermethylation as a molecular biomarker for prostate cancer. J Cell Biochem. 2004;91:540–552. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

