Activation of NRF2 by nitrosative agents and H2O2 involves KEAP1 disulfide formation
- PMID: 20061377
- PMCID: PMC2832995
- DOI: 10.1074/jbc.M109.051714
Activation of NRF2 by nitrosative agents and H2O2 involves KEAP1 disulfide formation
Abstract
The NRF2 transcription factor regulates a major environmental and oxidative stress response. NRF2 is itself negatively regulated by KEAP1, the adaptor of a Cul3-ubiquitin ligase complex that marks NRF2 for proteasomal degradation by ubiquitination. Electrophilic compounds activate NRF2 primarily by inhibiting KEAP1-dependent NRF2 degradation, through alkylation of specific cysteines. We have examined the impact on KEAP1 of reactive oxygen and nitrogen species, which are also NRF2 inducers. We found that in untreated cells, a fraction of KEAP1 carried a long range disulfide linking Cys(226) and Cys(613). Exposing cells to hydrogen peroxide, to the nitric oxide donor spermine NONOate, to hypochlorous acid, or to S-nitrosocysteine further increased this disulfide and promoted formation of a disulfide linking two KEAP1 molecules via Cys(151). None of these oxidants, except S-nitrocysteine, caused KEAP1 S-nitrosylation. A cysteine mutant preventing KEAP1 intermolecular disulfide formation also prevented NRF2 stabilization in response to oxidants, whereas those preventing intramolecular disulfide formation were functionally silent. Further, simultaneously inactivating the thioredoxin and glutathione pathways led both to major constitutive KEAP1 oxidation and NRF2 stabilization. We propose that KEAP1 intermolecular disulfide formation via Cys(151) underlies the activation of NRF2 by reactive oxygen and nitrogen species.
Figures



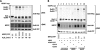

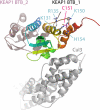
Similar articles
-
The gasotransmitter hydrogen sulfide induces nrf2-target genes by inactivating the keap1 ubiquitin ligase substrate adaptor through formation of a disulfide bond between cys-226 and cys-613.Antioxid Redox Signal. 2013 Aug 10;19(5):465-81. doi: 10.1089/ars.2012.4944. Epub 2013 Jan 16. Antioxid Redox Signal. 2013. PMID: 23145493
-
The Keap1-Nrf2 system as an in vivo sensor for electrophiles.Nitric Oxide. 2011 Aug 1;25(2):153-60. doi: 10.1016/j.niox.2011.02.007. Epub 2011 Mar 6. Nitric Oxide. 2011. PMID: 21385624
-
Signaling pathways activated by the phytochemical nordihydroguaiaretic acid contribute to a Keap1-independent regulation of Nrf2 stability: Role of glycogen synthase kinase-3.Free Radic Biol Med. 2012 Jan 15;52(2):473-87. doi: 10.1016/j.freeradbiomed.2011.11.003. Epub 2011 Nov 12. Free Radic Biol Med. 2012. PMID: 22142471
-
Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution.Genes Cells. 2011 Feb;16(2):123-40. doi: 10.1111/j.1365-2443.2010.01473.x. Genes Cells. 2011. PMID: 21251164 Review.
-
Interplay between cytosolic disulfide reductase systems and the Nrf2/Keap1 pathway.Biochem Soc Trans. 2015 Aug;43(4):632-8. doi: 10.1042/BST20150021. Epub 2015 Aug 3. Biochem Soc Trans. 2015. PMID: 26551704 Free PMC article. Review.
Cited by
-
Oxidative stress: An essential factor in the process of arteriovenous fistula failure.Front Cardiovasc Med. 2022 Aug 11;9:984472. doi: 10.3389/fcvm.2022.984472. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 36035909 Free PMC article. Review.
-
Loss of SWI/SNF Chromatin Remodeling Alters NRF2 Signaling in Non-Small Cell Lung Carcinoma.Mol Cancer Res. 2020 Dec;18(12):1777-1788. doi: 10.1158/1541-7786.MCR-20-0082. Epub 2020 Aug 27. Mol Cancer Res. 2020. PMID: 32855269 Free PMC article.
-
Quinone-induced activation of Keap1/Nrf2 signaling by aspirin prodrugs masquerading as nitric oxide.Chem Res Toxicol. 2012 Dec 17;25(12):2725-36. doi: 10.1021/tx3003609. Epub 2012 Oct 18. Chem Res Toxicol. 2012. PMID: 23035985 Free PMC article.
-
Effects of Allyl Isothiocyanate on Oxidative and Inflammatory Stress in Type 2 Diabetic Rats.Molecules. 2022 Aug 29;27(17):5568. doi: 10.3390/molecules27175568. Molecules. 2022. PMID: 36080332 Free PMC article.
-
Homocysteine causes vascular endothelial dysfunction by disrupting endoplasmic reticulum redox homeostasis.Redox Biol. 2019 Jan;20:46-59. doi: 10.1016/j.redox.2018.09.021. Epub 2018 Sep 26. Redox Biol. 2019. PMID: 30292945 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

