A Drosophila insulin-like peptide promotes growth during nonfeeding states
- PMID: 20059956
- PMCID: PMC2806523
- DOI: 10.1016/j.devcel.2009.10.009
A Drosophila insulin-like peptide promotes growth during nonfeeding states
Abstract
In metazoans, tissue growth relies on the availability of nutrients--stored internally or obtained from the environment--and the resulting activation of insulin/IGF signaling (IIS). In Drosophila, growth is mediated by seven Drosophila insulin-like peptides (Dilps), acting through a canonical IIS pathway. During the larval period, animals feed and Dilps produced by the brain couple nutrient uptake with systemic growth. We show here that, during metamorphosis, when feeding stops, a specific DILP (Dilp6) is produced by the fat body and relays the growth signal. Expression of DILP6 during pupal development is controlled by the steroid hormone ecdysone. Remarkably, DILP6 expression is also induced upon starvation, and both its developmental and environmental expression require the Drosophila FoxO transcription factor. This study reveals a specific class of ILPs induced upon metabolic stress that promotes growth in conditions of nutritional deprivation or following developmentally induced cessation of feeding.
2009 Elsevier Inc. All rights reserved.
Figures
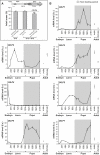
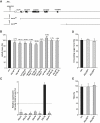
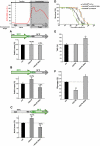
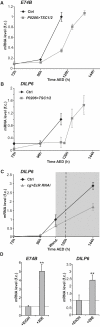
Similar articles
-
Drosophila insulin-like peptide-6 (dilp6) expression from fat body extends lifespan and represses secretion of Drosophila insulin-like peptide-2 from the brain.Aging Cell. 2012 Dec;11(6):978-85. doi: 10.1111/acel.12000. Epub 2012 Sep 18. Aging Cell. 2012. PMID: 22935001 Free PMC article.
-
A local insulin reservoir in Drosophila alpha cell homologs ensures developmental progression under nutrient shortage.Curr Biol. 2022 Apr 25;32(8):1788-1797.e5. doi: 10.1016/j.cub.2022.02.068. Epub 2022 Mar 21. Curr Biol. 2022. PMID: 35316653
-
Local requirement of the Drosophila insulin binding protein imp-L2 in coordinating developmental progression with nutritional conditions.Dev Biol. 2013 Sep 1;381(1):97-106. doi: 10.1016/j.ydbio.2013.06.008. Epub 2013 Jun 15. Dev Biol. 2013. PMID: 23773803
-
Insulin/IGF signaling in Drosophila and other insects: factors that regulate production, release and post-release action of the insulin-like peptides.Cell Mol Life Sci. 2016 Jan;73(2):271-90. doi: 10.1007/s00018-015-2063-3. Epub 2015 Oct 15. Cell Mol Life Sci. 2016. PMID: 26472340 Free PMC article. Review.
-
Nutrient availability and growth: regulation of insulin signaling by dFOXO/FOXO1.Cell Cycle. 2006 Mar;5(5):503-5. doi: 10.4161/cc.5.5.2501. Epub 2006 Mar 1. Cell Cycle. 2006. PMID: 16552183 Review.
Cited by
-
dj-1β regulates oxidative stress, insulin-like signaling and development in Drosophila melanogaster.Cell Cycle. 2012 Oct 15;11(20):3876-86. doi: 10.4161/cc.22073. Epub 2012 Sep 14. Cell Cycle. 2012. PMID: 22983063 Free PMC article.
-
Insulin signaling misregulation underlies circadian and cognitive deficits in a Drosophila fragile X model.Mol Psychiatry. 2017 Aug;22(8):1140-1148. doi: 10.1038/mp.2016.51. Epub 2016 Apr 19. Mol Psychiatry. 2017. PMID: 27090306 Free PMC article.
-
Insulin signaling regulates neurite growth during metamorphic neuronal remodeling.Biol Open. 2014 Jan 15;3(1):81-93. doi: 10.1242/bio.20136437. Biol Open. 2014. PMID: 24357229 Free PMC article.
-
Feeding-State-Dependent Modulation of Temperature Preference Requires Insulin Signaling in Drosophila Warm-Sensing Neurons.Curr Biol. 2018 Mar 5;28(5):779-787.e3. doi: 10.1016/j.cub.2018.01.060. Epub 2018 Feb 22. Curr Biol. 2018. PMID: 29478858 Free PMC article.
-
Regulation of adult stem cell behavior by nutrient signaling.Cell Cycle. 2011 Aug 15;10(16):2628-34. doi: 10.4161/cc.10.16.17059. Epub 2011 Aug 15. Cell Cycle. 2011. PMID: 21814033 Free PMC article.
References
-
- Aguila J.R., Suszko J., Gibbs A.G., Hoshizaki D.K. The role of larval fat cells in adult Drosophila melanogaster. J. Exp. Biol. 2007;210:956–963. - PubMed
-
- Britton J.S., Edgar B.A. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;125:2149–2158. - PubMed
-
- Brogiolo W., Stocker H., Ikeya T., Rintelen F., Fernandez R., Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 2001;11:213–221. - PubMed
-
- Broughton S.J., Piper M.D., Ikeya T., Bass T.M., Jacobson J., Driege Y., Martinez P., Hafen E., Withers D.J., Leevers S.J. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. USA. 2005;102:3105–3110. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

