Adaptation of Hansenula polymorpha to methanol: a transcriptome analysis
- PMID: 20044946
- PMCID: PMC2827406
- DOI: 10.1186/1471-2164-11-1
Adaptation of Hansenula polymorpha to methanol: a transcriptome analysis
Abstract
Background: Methylotrophic yeast species (e.g. Hansenula polymorpha, Pichia pastoris) can grow on methanol as sole source of carbon and energy. These organisms are important cell factories for the production of recombinant proteins, but are also used in fundamental research as model organisms to study peroxisome biology. During exponential growth on glucose, cells of H. polymorpha typically contain a single, small peroxisome that is redundant for growth while on methanol multiple, enlarged peroxisomes are present. These organelles are crucial to support growth on methanol, as they contain key enzymes of methanol metabolism.In this study, changes in the transcriptional profiles during adaptation of H. polymorpha cells from glucose- to methanol-containing media were investigated using DNA-microarray analyses.
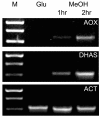
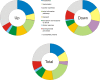
Results: Two hours after the shift of cells from glucose to methanol nearly 20% (1184 genes) of the approximately 6000 annotated H. polymorpha genes were significantly upregulated with at least a two-fold differential expression. Highest upregulation (> 300-fold) was observed for the genes encoding the transcription factor Mpp1 and formate dehydrogenase, an enzyme of the methanol dissimilation pathway. Upregulated genes also included genes encoding other enzymes of methanol metabolism as well as of peroxisomal beta-oxidation.A moderate increase in transcriptional levels (up to 4-fold) was observed for several PEX genes, which are involved in peroxisome biogenesis. Only PEX11 and PEX32 were higher upregulated. In addition, an increase was observed in expression of the several ATG genes, which encode proteins involved in autophagy and autophagy processes. The strongest upregulation was observed for ATG8 and ATG11.Approximately 20% (1246 genes) of the genes were downregulated. These included glycolytic genes as well as genes involved in transcription and translation.
Conclusion: Transcriptional profiling of H. polymorpha cells shifted from glucose to methanol showed the expected downregulation of glycolytic genes together with upregulation of the methanol utilisation pathway. This serves as a confirmation and validation of the array data obtained. Consistent with this, also various PEX genes were upregulated. The strong upregulation of ATG genes is possibly due to induction of autophagy processes related to remodeling of the cell architecture required to support growth on methanol. These processes may also be responsible for the enhanced peroxisomal beta-oxidation, as autophagy leads to recycling of membrane lipids. The prominent downregulation of transcription and translation may be explained by the reduced growth rate on methanol (td glucose 1 h vs td methanol 4.5 h).
Figures


Similar articles
-
Genome sequence and analysis of methylotrophic yeast Hansenula polymorpha DL1.BMC Genomics. 2013 Nov 27;14:837. doi: 10.1186/1471-2164-14-837. BMC Genomics. 2013. PMID: 24279325 Free PMC article.
-
Transcriptome analysis of Δmig1Δmig2 mutant reveals their roles in methanol catabolism, peroxisome biogenesis and autophagy in methylotrophic yeast Pichia pastoris.Genes Genomics. 2018 Apr;40(4):399-412. doi: 10.1007/s13258-017-0641-5. Epub 2017 Dec 14. Genes Genomics. 2018. PMID: 29892842
-
Hansenula polymorpha Tup1p is important for peroxisome degradation.FEMS Yeast Res. 2004 Sep;4(8):789-94. doi: 10.1016/j.femsyr.2004.04.006. FEMS Yeast Res. 2004. PMID: 15450185
-
Pexophagy: the selective autophagy of peroxisomes.Autophagy. 2005 Jul;1(2):75-83. doi: 10.4161/auto.1.2.1737. Epub 2005 Jul 13. Autophagy. 2005. PMID: 16874024 Review.
-
Peroxisome homeostasis in Hansenula polymorpha.FEMS Yeast Res. 2003 Nov;4(2):131-9. doi: 10.1016/S1567-1356(03)00070-9. FEMS Yeast Res. 2003. PMID: 14613877 Review.
Cited by
-
STED super-resolution microscopy unveils the dynamics of Atg30 on yeast Pex3-labeled peroxisomes.iScience. 2024 Jul 8;27(8):110481. doi: 10.1016/j.isci.2024.110481. eCollection 2024 Aug 16. iScience. 2024. PMID: 39156652 Free PMC article.
-
Atg8: an autophagy-related ubiquitin-like protein family.Genome Biol. 2011 Jul 27;12(7):226. doi: 10.1186/gb-2011-12-7-226. Genome Biol. 2011. PMID: 21867568 Free PMC article. Review.
-
Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1.Autophagy. 2021 Jan;17(1):1-382. doi: 10.1080/15548627.2020.1797280. Epub 2021 Feb 8. Autophagy. 2021. PMID: 33634751 Free PMC article.
-
Genome evolution and transcriptome plasticity is associated with adaptation to monocot and dicot plants in Colletotrichum fungi.Gigascience. 2024 Jan 2;13:giae036. doi: 10.1093/gigascience/giae036. Gigascience. 2024. PMID: 38940768 Free PMC article.
-
Post-translational modifications of proteins associated with yeast peroxisome membrane: An essential mode of regulatory mechanism.Genes Cells. 2021 Nov;26(11):843-860. doi: 10.1111/gtc.12892. Epub 2021 Sep 2. Genes Cells. 2021. PMID: 34472666 Free PMC article. Review.
References
-
- Gellissen G, Kunze G, Gaillardin C, Cregg JM, Berardi E, Veenhuis M, Klei IJ van der. New yeast expression platforms based on methylotrophic Hansenula polymorpha and Pichia pastoris and on dimorphic Arxula Adeninivorans and Yarrowia lipolytica - A comparison. FEMS Yeast Res. 2005;5:1079–1096. doi: 10.1016/j.femsyr.2005.06.004. - DOI - PubMed
-
- Veenhuis M, Keizer I, Harder W. Characterization of peroxisomes in glucosegrown Hansenula polymorpha and their development after the transfer of cells into methanol-containing media. Arch Microbiol. 1979;120:167–175. doi: 10.1007/BF00409104. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

