Evolution of the HIV-1 env gene in the Rag2-/- gammaC-/- humanized mouse model
- PMID: 20042504
- PMCID: PMC2826074
- DOI: 10.1128/JVI.02180-09
Evolution of the HIV-1 env gene in the Rag2-/- gammaC-/- humanized mouse model
Abstract
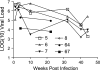
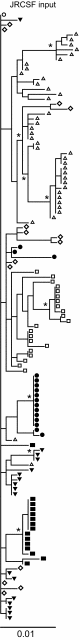
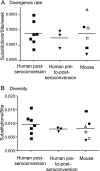
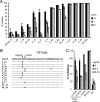
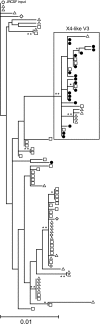
Rag2(-/-) gamma(C)(-/-) mice transplanted with human hematopoietic stem cells (DKO-hu-HSC mice) mimic aspects of human infection with human immunodeficiency virus type 1 (HIV-1), including sustained viral replication and CD4(+) T-cell decline. However, the extent of HIV-1 evolution during long-term infection in these humanized mice, a key feature of the natural infection, has not been assessed fully. In this study, we examined the types of genotypic and phenotypic changes in the viral env gene that occur in the viral populations of DKO-hu-HSC mice infected with the CCR5-tropic isolate HIV-1(JRCSF) for up to 44 weeks. The mean rate of divergence of viral populations in mice was similar to that observed in a cohort of humans during a similar period of infection. Many amino acid substitutions were common across mice, including losses of N-linked glycosylation sites and substitutions in the CD4 binding site and in CD4-induced epitopes, indicating common selective pressures between mice. In addition, env variants evolved sensitivity to antibodies directed at V3, suggesting a more open conformation for Env. This phenotypic change was associated with increased CD4 binding efficiency and was attributed to specific amino acid substitutions. In one mouse, env variants emerged that exhibited a CXCR4-tropic phenotype. These sequences were compartmentalized in the mesenteric lymph node. In summary, viral populations in these mice exhibited dynamic behavior that included sequence evolution, compartmentalization, and the appearance of distinct phenotypic changes. Thus, humanized mice offer a useful model for studying evolutionary processes of HIV-1 in a complex host environment.
Figures

Similar articles
-
Mucosal transmission of R5 and X4 tropic HIV-1 via vaginal and rectal routes in humanized Rag2-/- gammac -/- (RAG-hu) mice.Virology. 2008 Apr 10;373(2):342-51. doi: 10.1016/j.virol.2007.11.020. Epub 2008 Jan 18. Virology. 2008. PMID: 18207484 Free PMC article.
-
A comparative study of HIV-1 clade C env evolution in a Zambian infant with an infected rhesus macaque during disease progression.AIDS. 2009 Sep 10;23(14):1817-28. doi: 10.1097/QAD.0b013e32832f3da6. AIDS. 2009. PMID: 19609201 Free PMC article.
-
Experimental Adaptive Evolution of Simian Immunodeficiency Virus SIVcpz to Pandemic Human Immunodeficiency Virus Type 1 by Using a Humanized Mouse Model.J Virol. 2018 Jan 30;92(4):e01905-17. doi: 10.1128/JVI.01905-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29212937 Free PMC article.
-
Current humanized mouse models for studying human immunology and HIV-1 immuno-pathogenesis.Sci China Life Sci. 2010 Feb;53(2):195-203. doi: 10.1007/s11427-010-0059-7. Epub 2010 Mar 7. Sci China Life Sci. 2010. PMID: 20596827 Free PMC article. Review.
-
The utility of the new generation of humanized mice to study HIV-1 infection: transmission, prevention, pathogenesis, and treatment.Retrovirology. 2011 Aug 11;8:65. doi: 10.1186/1742-4690-8-65. Retrovirology. 2011. PMID: 21835012 Free PMC article. Review.
Cited by
-
New generation humanized mice for virus research: comparative aspects and future prospects.Virology. 2013 Jan 5;435(1):14-28. doi: 10.1016/j.virol.2012.10.007. Virology. 2013. PMID: 23217612 Free PMC article. Review.
-
HIV restriction by APOBEC3 in humanized mice.PLoS Pathog. 2013 Mar;9(3):e1003242. doi: 10.1371/journal.ppat.1003242. Epub 2013 Mar 28. PLoS Pathog. 2013. PMID: 23555255 Free PMC article.
-
Mice transgenic for CD4-specific human CD4, CCR5 and cyclin T1 expression: a new model for investigating HIV-1 transmission and treatment efficacy.PLoS One. 2013 May 15;8(5):e63537. doi: 10.1371/journal.pone.0063537. Print 2013. PLoS One. 2013. PMID: 23691059 Free PMC article.
-
HIV-2 genetic evolution in patients with advanced disease is faster than that in matched HIV-1 patients.J Virol. 2010 Jul;84(14):7412-5. doi: 10.1128/JVI.02548-09. Epub 2010 May 12. J Virol. 2010. PMID: 20463072 Free PMC article.
-
Remarkable lethal G-to-A mutations in vif-proficient HIV-1 provirus by individual APOBEC3 proteins in humanized mice.J Virol. 2010 Sep;84(18):9546-56. doi: 10.1128/JVI.00823-10. Epub 2010 Jul 7. J Virol. 2010. PMID: 20610708 Free PMC article.
References
-
- Baba, M., O. Nishimura, N. Kanzaki, M. Okamoto, H. Sawada, Y. Iizawa, M. Shiraishi, Y. Aramaki, K. Okonogi, Y. Ogawa, K. Meguro, and M. Fujino. 1999. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. USA 96:5698-5703. - PMC - PubMed
-
- Baenziger, S., R. Tussiwand, E. Schlaepfer, L. Mazzucchelli, M. Heikenwalder, M. O. Kurrer, S. Behnke, J. Frey, A. Oxenius, H. Joller, A. Aguzzi, M. G. Manz, and R. F. Speck. 2006. Disseminated and sustained HIV infection in CD34+ cord blood cell-transplanted Rag2−/−gamma c−/− mice. Proc. Natl. Acad. Sci. USA 103:15951-15956. - PMC - PubMed
-
- Beaumont, T., E. Quakkelaar, A. van Nuenen, R. Pantophlet, and H. Schuitemaker. 2004. Increased sensitivity to CD4 binding site-directed neutralization following in vitro propagation on primary lymphocytes of a neutralization-resistant human immunodeficiency virus IIIB strain isolated from an accidentally infected laboratory worker. J. Virol. 78:5651-5657. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

