Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library
- PMID: 20037582
- PMCID: PMC2951730
- DOI: 10.1038/nbt.1600
Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library
Abstract
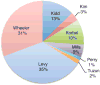
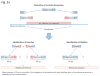
Structural variants (SVs) are a major source of human genomic variation; however, characterizing them at nucleotide resolution remains challenging. Here we assemble a library of breakpoints at nucleotide resolution from collating and standardizing ~2,000 published SVs. For each breakpoint, we infer its ancestral state (through comparison to primate genomes) and its mechanism of formation (e.g., nonallelic homologous recombination, NAHR). We characterize breakpoint sequences with respect to genomic landmarks, chromosomal location, sequence motifs and physical properties, finding that the occurrence of insertions and deletions is more balanced than previously reported and that NAHR-formed breakpoints are associated with relatively rigid, stable DNA helices. Finally, we demonstrate an approach, BreakSeq, for scanning the reads from short-read sequenced genomes against our breakpoint library to accurately identify previously overlooked SVs, which we then validate by PCR. As new data become available, we expect our BreakSeq approach will become more sensitive and facilitate rapid SV genotyping of personal genomes.
Figures


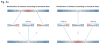
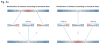








Similar articles
-
Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome.Genome Res. 2010 May;20(5):623-35. doi: 10.1101/gr.102970.109. Epub 2010 Mar 22. Genome Res. 2010. PMID: 20308636 Free PMC article.
-
Small polymorphisms are a source of ancestral bias in structural variant breakpoint placement.Genome Res. 2024 Feb 7;34(1):7-19. doi: 10.1101/gr.278203.123. Genome Res. 2024. PMID: 38176712 Free PMC article.
-
The fine-scale architecture of structural variants in 17 mouse genomes.Genome Biol. 2012;13(3):R18. doi: 10.1186/gb-2012-13-3-r18. Genome Biol. 2012. PMID: 22439878 Free PMC article.
-
Challenges in studying genomic structural variant formation mechanisms: the short-read dilemma and beyond.Bioessays. 2011 Nov;33(11):840-50. doi: 10.1002/bies.201100075. Epub 2011 Sep 30. Bioessays. 2011. PMID: 21959584 Review.
-
Geographic distribution and adaptive significance of genomic structural variants: an anthropological genetics perspective.Hum Biol. 2014 Fall;86(4):260-75. doi: 10.13110/humanbiology.86.4.0260. Hum Biol. 2014. PMID: 25959693 Review.
Cited by
-
An improved understanding of cancer genomics through massively parallel sequencing.Transl Cancer Res. 2014 Jun;3(3):243-259. doi: 10.3978/j.issn.2218-676X.2014.05.05. Transl Cancer Res. 2014. PMID: 26146607 Free PMC article.
-
Characteristics of de novo structural changes in the human genome.Genome Res. 2015 Jun;25(6):792-801. doi: 10.1101/gr.185041.114. Epub 2015 Apr 16. Genome Res. 2015. PMID: 25883321 Free PMC article.
-
Novel variation and de novo mutation rates in population-wide de novo assembled Danish trios.Nat Commun. 2015 Jan 19;6:5969. doi: 10.1038/ncomms6969. Nat Commun. 2015. PMID: 25597990 Free PMC article.
-
A map of human genome variation from population-scale sequencing.Nature. 2010 Oct 28;467(7319):1061-73. doi: 10.1038/nature09534. Nature. 2010. PMID: 20981092 Free PMC article.
-
Amplification and thrifty single-molecule sequencing of recurrent somatic structural variations.Genome Res. 2014 Feb;24(2):318-28. doi: 10.1101/gr.161497.113. Epub 2013 Dec 4. Genome Res. 2014. PMID: 24307551 Free PMC article.
References
-
- Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. - PubMed
-
- Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. - PubMed
-
- Tuzun E, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–732. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

