A human cytomegalovirus gO-null mutant fails to incorporate gH/gL into the virion envelope and is unable to enter fibroblasts and epithelial and endothelial cells
- PMID: 20032184
- PMCID: PMC2820920
- DOI: 10.1128/JVI.02249-09
A human cytomegalovirus gO-null mutant fails to incorporate gH/gL into the virion envelope and is unable to enter fibroblasts and epithelial and endothelial cells
Abstract
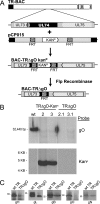
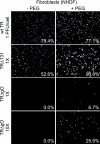
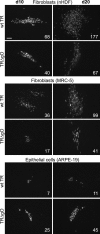
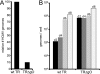
Human cytomegalovirus (HCMV) depends upon a five-protein complex, gH/gL/UL128-131, to enter epithelial and endothelial cells. A separate HCMV gH/gL-containing complex, gH/gL/gO, has been described. Our prevailing model is that gH/gL/UL128-131 is required for entry into biologically important epithelial and endothelial cells and that gH/gL/gO is required for infection of fibroblasts. Genes encoding UL128-131 are rapidly mutated during laboratory propagation of HCMV on fibroblasts, apparently related to selective pressure for the fibroblast entry pathway. Arguing against this model in the accompanying paper by B. J. Ryckman et al. (J. Virol., 84:2597-2609, 2010), we describe evidence that clinical HCMV strain TR expresses a gO molecule that acts to promote endoplasmic reticulum (ER) export of gH/gL and that gO is not stably incorporated into the virus envelope. This was different from results involving fibroblast-adapted HCMV strain AD169, which incorporates gO into the virion envelope. Here, we constructed a TR gO-null mutant, TRDeltagO, that replicated to low titers, spread poorly among fibroblasts, but produced normal quantities of extracellular virus particles. TRDeltagO particles released from fibroblasts failed to infect fibroblasts and epithelial and endothelial cells, but the chemical fusogen polyethylene glycol (PEG) could partially overcome defects in infection. Therefore, TRDeltagO is defective for entry into all three cell types. Defects in entry were explained by observations showing that TRDeltagO incorporated about 5% of the quantities of gH/gL in extracellular virus particles compared with that in wild-type virions. Although TRDeltagO particles could not enter cells, cell-to-cell spread involving epithelial and endothelial cells was increased relative to TR, apparently resulting from increased quantities of gH/gL/UL128-131 in virions. Together, our data suggest that TR gO acts as a chaperone to promote ER export and the incorporation of gH/gL complexes into the HCMV envelope. Moreover, these data suggest that it is gH/gL, and not gH/gL/gO, that is present in virions and is required for infection of fibroblasts and epithelial and endothelial cells. Our observations that both gH/gL and gH/gL/UL128-131 are required for entry into epithelial/endothelial cells differ from models for other beta- and gammaherpesviruses that use one of two different gH/gL complexes to enter different cells.
Figures


Similar articles
-
Human cytomegalovirus TR strain glycoprotein O acts as a chaperone promoting gH/gL incorporation into virions but is not present in virions.J Virol. 2010 Mar;84(5):2597-609. doi: 10.1128/JVI.02256-09. Epub 2009 Dec 23. J Virol. 2010. PMID: 20032193 Free PMC article.
-
Human Cytomegalovirus gH/gL/gO Promotes the Fusion Step of Entry into All Cell Types, whereas gH/gL/UL128-131 Broadens Virus Tropism through a Distinct Mechanism.J Virol. 2015 Sep;89(17):8999-9009. doi: 10.1128/JVI.01325-15. Epub 2015 Jun 17. J Virol. 2015. PMID: 26085146 Free PMC article.
-
Loss of the Human Cytomegalovirus US16 Protein Abrogates Virus Entry into Endothelial and Epithelial Cells by Reducing the Virion Content of the Pentamer.J Virol. 2017 May 12;91(11):e00205-17. doi: 10.1128/JVI.00205-17. Print 2017 Jun 1. J Virol. 2017. PMID: 28331097 Free PMC article.
-
Human cytomegalovirus tropism for endothelial/epithelial cells: scientific background and clinical implications.Rev Med Virol. 2010 May;20(3):136-55. doi: 10.1002/rmv.645. Rev Med Virol. 2010. PMID: 20084641 Review.
-
Principles for studying in vivo attenuation of virus mutants: defining the role of the cytomegalovirus gH/gL/gO complex as a paradigm.Med Microbiol Immunol. 2015 Jun;204(3):295-305. doi: 10.1007/s00430-015-0405-2. Epub 2015 Mar 18. Med Microbiol Immunol. 2015. PMID: 25782576 Review.
Cited by
-
Human Cytomegalovirus Host Interactions: EGFR and Host Cell Signaling Is a Point of Convergence Between Viral Infection and Functional Changes in Infected Cells.Front Microbiol. 2021 May 7;12:660901. doi: 10.3389/fmicb.2021.660901. eCollection 2021. Front Microbiol. 2021. PMID: 34025614 Free PMC article. Review.
-
Antibodies against the gH/gL/UL128/UL130/UL131 complex comprise the majority of the anti-cytomegalovirus (anti-CMV) neutralizing antibody response in CMV hyperimmune globulin.J Virol. 2012 Jul;86(13):7444-7. doi: 10.1128/JVI.00467-12. Epub 2012 Apr 24. J Virol. 2012. PMID: 22532696 Free PMC article.
-
Cytomegalovirus vaccines and methods of production (WO20009049138): the emerging recognition of the importance of virus neutralization at the epithelial/endothelial interface.Expert Opin Ther Pat. 2010 Apr;20(4):597-602. doi: 10.1517/13543770903584882. Expert Opin Ther Pat. 2010. PMID: 20302454 Free PMC article.
-
Virus-Like Particles and Nanoparticles for Vaccine Development against HCMV.Viruses. 2019 Dec 28;12(1):35. doi: 10.3390/v12010035. Viruses. 2019. PMID: 31905677 Free PMC article. Review.
-
The transcriptome of human mammary epithelial cells infected with the HCMV-DB strain displays oncogenic traits.Sci Rep. 2018 Aug 22;8(1):12574. doi: 10.1038/s41598-018-30109-1. Sci Rep. 2018. PMID: 30135434 Free PMC article.
References
-
- Adler, B., L. Scrivano, Z. Ruzcics, B. Rupp, C. Sinzger, and U. Koszinowski. 2006. Role of human cytomegalovirus UL131A in cell type-specific virus entry and release. J. Gen. Virol. 87:2451-2460. - PubMed
-
- Akter, P., C. Cunningham, B. P. McSharry, A. Dolan, C. Addison, D. J. Dargan, A. F. Hassan-Walker, V. C. Emery, P. D. Griffiths, G. W. Wilkinson, and A. J. Davison. 2003. Two novel spliced genes in human cytomegalovirus. J. Gen. Virol. 84:1117-1122. - PubMed
-
- Bissinger, A. L., C. Sinzger, E. Kaiserling, and G. Jahn. 2002. Human cytomegalovirus as a direct pathogen: correlation of multiorgan involvement and cell distribution with clinical and pathological findings in a case of congenital inclusion disease. J. Med. Virol. 67:200-206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

