RNF4 and VHL regulate the proteasomal degradation of SUMO-conjugated Hypoxia-Inducible Factor-2alpha
- PMID: 20026589
- PMCID: PMC2847224
- DOI: 10.1093/nar/gkp1157
RNF4 and VHL regulate the proteasomal degradation of SUMO-conjugated Hypoxia-Inducible Factor-2alpha
Abstract
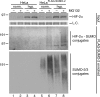
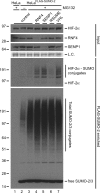
Hypoxia-inducible factors (HIFs) are critical transcription factors that mediate cell survival during reduced oxygen conditions (hypoxia). At regular oxygen conditions (normoxia), HIF-1alpha and HIF-2alpha are continuously synthesized in cells and degraded via the ubiquitin-proteasome pathway. During hypoxia, these proteins are stabilized and translocate to the nucleus to activate transcription of target genes that enable cell survival at reduced oxygen levels. HIF proteins are tightly regulated via post-translational modifications including phosphorylation, acetylation, prolyl-hydroxylation and ubiquitination. Here we show for the first time that exogenous and endogenous HIF-2alpha are also regulated via the ubiquitin-like modifier small ubiquitin-like modifiers (SUMO). Using mutational analysis, we found that K394, which is situated in the sumoylation consensus site LKEE, is the major SUMO acceptor site in HIF-2alpha. Functionally, sumoylation reduced the transcriptional activity of HIF-2alpha. Similar to HIF-1alpha, HIF-2alpha is regulated by the SUMO protease SENP1. The proteasome inhibitor MG132 strongly stabilized SUMO-2-conjugated HIF-2alpha during hypoxia but did not affect the total level of HIF-2alpha. The ubiquitin E3 ligases von Hippel-Lindau and RNF4 control the levels of sumoylated HIF-2alpha, indicating that sumoylated HIF-2alpha is degraded via SUMO-targeted ubiquitin ligases.
Figures

Similar articles
-
c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4.Cell Cycle. 2015;14(12):1859-72. doi: 10.1080/15384101.2015.1040965. Cell Cycle. 2015. PMID: 25895136 Free PMC article.
-
Up-regulation of hypoxia-inducible factors HIF-1alpha and HIF-2alpha under normoxic conditions in renal carcinoma cells by von Hippel-Lindau tumor suppressor gene loss of function.Oncogene. 2000 Nov 16;19(48):5435-43. doi: 10.1038/sj.onc.1203938. Oncogene. 2000. PMID: 11114720
-
Flavonoids-induced accumulation of hypoxia-inducible factor (HIF)-1alpha/2alpha is mediated through chelation of iron.J Cell Biochem. 2008 Apr 15;103(6):1989-98. doi: 10.1002/jcb.21588. J Cell Biochem. 2008. PMID: 17973296
-
Regulation of HIF by the von Hippel-Lindau tumour suppressor: implications for cellular oxygen sensing.IUBMB Life. 2001 Jul;52(1-2):43-7. doi: 10.1080/15216540252774757. IUBMB Life. 2001. PMID: 11795592 Review.
-
Von Hippel-Lindau tumor suppressor protein and hypoxia-inducible factor in kidney cancer.Am J Nephrol. 2004 Jan-Feb;24(1):1-13. doi: 10.1159/000075346. Epub 2003 Dec 3. Am J Nephrol. 2004. PMID: 14654728 Review.
Cited by
-
A SUMOylation-dependent HIF-1α/CLDN6 negative feedback mitigates hypoxia-induced breast cancer metastasis.J Exp Clin Cancer Res. 2020 Feb 24;39(1):42. doi: 10.1186/s13046-020-01547-5. J Exp Clin Cancer Res. 2020. PMID: 32093760 Free PMC article.
-
Astragaloside IV protects cardiomyocytes against hypoxia injury via HIF-1α and the JAK2/STAT3 pathway.Ann Transl Med. 2021 Sep;9(18):1435. doi: 10.21037/atm-21-4080. Ann Transl Med. 2021. PMID: 34733987 Free PMC article.
-
Centromere binding and a conserved role in chromosome stability for SUMO-dependent ubiquitin ligases.PLoS One. 2013 Jun 13;8(6):e65628. doi: 10.1371/journal.pone.0065628. Print 2013. PLoS One. 2013. PMID: 23785440 Free PMC article.
-
The SUMOylation and ubiquitination crosstalk in cancer.J Cancer Res Clin Oncol. 2023 Nov;149(17):16123-16146. doi: 10.1007/s00432-023-05310-z. Epub 2023 Aug 28. J Cancer Res Clin Oncol. 2023. PMID: 37640846 Review.
-
Expression of HIF-2α and VEGF in Cervical Squamous Cell Carcinoma and Its Clinical Significance.Biomed Res Int. 2016;2016:5631935. doi: 10.1155/2016/5631935. Epub 2016 Jun 20. Biomed Res Int. 2016. PMID: 27413748 Free PMC article. Clinical Trial.
References
-
- Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. - PubMed
-
- Meulmeester E, Melchior F. Cell biology: SUMO. Nature. 2008;452:709–711. - PubMed
-
- Chen A, Mannen H, Li SS. Characterization of mouse ubiquitin-like SMT3A and SMT3B cDNAs and gene/pseudogenes. Biochem. Mol. Biol. Int. 1998;46:1161–1174. - PubMed
-
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. - PubMed
-
- Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell Proteomics. 2006;5:2298–2310. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

