Regulation of mRNA cap methylation
- PMID: 20025612
- PMCID: PMC2825737
- DOI: 10.1042/BJ20091352
Regulation of mRNA cap methylation
Abstract
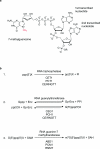
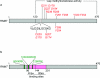
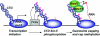
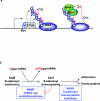
The 7-methylguanosine cap added to the 5' end of mRNA is essential for efficient gene expression and cell viability. Methylation of the guanosine cap is necessary for the translation of most cellular mRNAs in all eukaryotic organisms in which it has been investigated. In some experimental systems, cap methylation has also been demonstrated to promote transcription, splicing, polyadenylation and nuclear export of mRNA. The present review discusses how the 7-methylguanosine cap is synthesized by cellular enzymes, the impact that the 7-methylguanosine cap has on biological processes, and how the mRNA cap methylation reaction is regulated.
Figures
Similar articles
-
Cap-binding complex (CBC).Biochem J. 2014 Jan 15;457(2):231-42. doi: 10.1042/BJ20131214. Biochem J. 2014. PMID: 24354960 Free PMC article. Review.
-
RAM/Fam103a1 is required for mRNA cap methylation.Mol Cell. 2011 Nov 18;44(4):585-96. doi: 10.1016/j.molcel.2011.08.041. Mol Cell. 2011. PMID: 22099306 Free PMC article.
-
Identification of a quality-control mechanism for mRNA 5'-end capping.Nature. 2010 Sep 30;467(7315):608-11. doi: 10.1038/nature09338. Epub 2010 Aug 29. Nature. 2010. PMID: 20802481 Free PMC article.
-
Thermodynamics of 7-methylguanosine cation stacking with tryptophan upon mRNA 5' cap binding to translation factor eIF4E.Nucleosides Nucleotides Nucleic Acids. 2003 May-Aug;22(5-8):1557-61. doi: 10.1081/NCN-120023033. Nucleosides Nucleotides Nucleic Acids. 2003. PMID: 14565465
-
Cap and cap-binding proteins in the control of gene expression.Wiley Interdiscip Rev RNA. 2011 Mar-Apr;2(2):277-98. doi: 10.1002/wrna.52. Epub 2010 Oct 28. Wiley Interdiscip Rev RNA. 2011. PMID: 21957010 Review.
Cited by
-
Identification of N7-methylguanosine related signature for prognosis and immunotherapy efficacy prediction in lung adenocarcinoma.Front Med (Lausanne). 2022 Aug 24;9:962972. doi: 10.3389/fmed.2022.962972. eCollection 2022. Front Med (Lausanne). 2022. PMID: 36091687 Free PMC article.
-
The covalent nucleotide modifications within plant mRNAs: What we know, how we find them, and what should be done in the future.Plant Cell. 2023 May 29;35(6):1801-1816. doi: 10.1093/plcell/koad044. Plant Cell. 2023. PMID: 36794718 Free PMC article.
-
Human RNA cap1 methyltransferase CMTr1 cooperates with RNA helicase DHX15 to modify RNAs with highly structured 5' termini.Philos Trans R Soc Lond B Biol Sci. 2018 Nov 5;373(1762):20180161. doi: 10.1098/rstb.2018.0161. Philos Trans R Soc Lond B Biol Sci. 2018. PMID: 30397098 Free PMC article.
-
Internal m7G methylation: A novel epitranscriptomic contributor in brain development and diseases.Mol Ther Nucleic Acids. 2023 Jan 11;31:295-308. doi: 10.1016/j.omtn.2023.01.003. eCollection 2023 Mar 14. Mol Ther Nucleic Acids. 2023. PMID: 36726408 Free PMC article. Review.
-
m7G Methylation-Related Genes as Biomarkers for Predicting Overall Survival Outcomes for Hepatocellular Carcinoma.Front Bioeng Biotechnol. 2022 May 10;10:849756. doi: 10.3389/fbioe.2022.849756. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 35620469 Free PMC article.
References
-
- Shuman S. What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell. Biol. 2002;3:619–625. - PubMed
-
- Shatkin A. J. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. - PubMed
-
- Shibagaki Y., Itoh N., Yamada H., Nagata S., Mizumoto K. mRNA capping enzyme. Isolation and characterization of the gene encoding mRNA guanylytransferase subunit from Saccharomyces cerevisiae. J. Biol. Chem. 1992;267:9521–9528. - PubMed
-
- Tsukamoto T., Shibagaki Y., Imajoh-Ohmi S., Murakoshi T., Suzuki M., Nakamura A., Gotoh H., Mizumoto K. Isolation and characterization of the yeast mRNA capping enzyme beta subunit gene encoding RNA 5′-triphosphatase, which is essential for cell viability. Biochem. Biophys. Res. Commun. 1997;239:116–122. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

