A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation
- PMID: 20023299
- PMCID: PMC2871419
- DOI: 10.1074/mcp.M900485-MCP200
A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation
Abstract
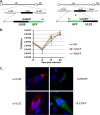
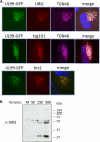
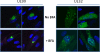
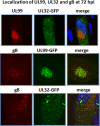
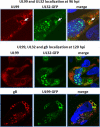
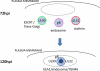
The assembly of infectious virus particles is a complex event. For human cytomegalovirus (HCMV) this process requires the coordinated expression and localization of at least 60 viral proteins that comprise the infectious virion. To gain insight into the mechanisms controlling this process, we identified protein binding partners for two viral proteins, pUL99 (also termed pp28) and pUL32 (pp150), which are essential for HCMV virion assembly. We utilized HCMV strains expressing pUL99 or pUL32 carboxyl-terminal green fluorescent protein fusion proteins from their native location in the HCMV genome. Based on the presence of ubiquitin in the pUL99 immunoisolation, we discovered that this viral protein colocalizes with components of the cellular endosomal sorting complex required for transport (ESCRT) pathway during the initial stages of virion assembly. We identified the nucleocapsid and a large number of tegument proteins as pUL32 binding partners, suggesting that events controlling trafficking of this viral protein in the cytoplasm regulate nucleocapsid/tegument maturation. The finding that pUL32, but not pUL99, associates with clathrin led to the discovery that the two viral proteins traffic via distinct pathways during the early stages of virion assembly. Additional investigation revealed that the majority of the major viral glycoprotein gB initially resides in a third compartment. Analysis of the trafficking of these three viral proteins throughout a time course of virion assembly allowed us to visualize their merger into a single large cytoplasmic structure during the late stages of viral assembly. We propose a model of HCMV virion maturation in which multiple components of the virion traffic independently of one another before merging.
Figures

Similar articles
-
Role of human cytomegalovirus tegument proteins in virion assembly.Viruses. 2014 Feb 6;6(2):582-605. doi: 10.3390/v6020582. Viruses. 2014. PMID: 24509811 Free PMC article. Review.
-
Potent Inhibition of Human Cytomegalovirus by Modulation of Cellular SNARE Syntaxin 5.J Virol. 2016 Dec 16;91(1):e01637-16. doi: 10.1128/JVI.01637-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795424 Free PMC article.
-
Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly.J Virol. 2000 Jan;74(2):975-86. doi: 10.1128/jvi.74.2.975-986.2000. J Virol. 2000. PMID: 10623760 Free PMC article.
-
Interaction of Human Cytomegalovirus Tegument Proteins ppUL35 and ppUL35A with Sorting Nexin 5 Regulates Glycoprotein B (gpUL55) Localization.J Virol. 2018 Apr 13;92(9):e00013-18. doi: 10.1128/JVI.00013-18. Print 2018 May 1. J Virol. 2018. PMID: 29444945 Free PMC article.
-
Human cytomegalovirus tegument proteins (pp65, pp71, pp150, pp28).Virol J. 2012 Jan 17;9:22. doi: 10.1186/1743-422X-9-22. Virol J. 2012. PMID: 22251420 Free PMC article. Review.
Cited by
-
Tegument Protein pp150 Sequence-Specific Peptide Blocks Cytomegalovirus Infection.Viruses. 2021 Nov 15;13(11):2277. doi: 10.3390/v13112277. Viruses. 2021. PMID: 34835083 Free PMC article.
-
Role of human cytomegalovirus tegument proteins in virion assembly.Viruses. 2014 Feb 6;6(2):582-605. doi: 10.3390/v6020582. Viruses. 2014. PMID: 24509811 Free PMC article. Review.
-
Manipulation of host pathways by human cytomegalovirus: insights from genome-wide studies.Semin Immunopathol. 2014 Nov;36(6):651-8. doi: 10.1007/s00281-014-0443-7. Epub 2014 Sep 27. Semin Immunopathol. 2014. PMID: 25260940 Review.
-
Proteomics-based methods for discovery, quantification, and validation of protein-protein interactions.Anal Chem. 2013 Jan 15;85(2):749-68. doi: 10.1021/ac3033257. Epub 2012 Dec 12. Anal Chem. 2013. PMID: 23157382 Free PMC article. Review. No abstract available.
-
The HCMV Assembly Compartment Is a Dynamic Golgi-Derived MTOC that Controls Nuclear Rotation and Virus Spread.Dev Cell. 2018 Apr 9;45(1):83-100.e7. doi: 10.1016/j.devcel.2018.03.010. Dev Cell. 2018. PMID: 29634939 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

