Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy
- PMID: 20019802
- PMCID: PMC2787017
- DOI: 10.1371/journal.pgen.1000773
Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy
Abstract
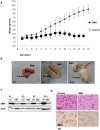
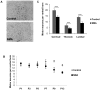
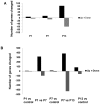
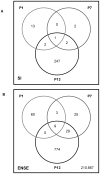
Spinal muscular atrophy is a severe motor neuron disease caused by inactivating mutations in the SMN1 gene leading to reduced levels of full-length functional SMN protein. SMN is a critical mediator of spliceosomal protein assembly, and complete loss or drastic reduction in protein leads to loss of cell viability. However, the reason for selective motor neuron degeneration when SMN is reduced to levels which are tolerated by all other cell types is not currently understood. Widespread splicing abnormalities have recently been reported at end-stage in a mouse model of SMA, leading to the proposition that disruption of efficient splicing is the primary mechanism of motor neuron death. However, it remains unclear whether splicing abnormalities are present during early stages of the disease, which would be a requirement for a direct role in disease pathogenesis. We performed exon-array analysis of RNA from SMN deficient mouse spinal cord at 3 time points, pre-symptomatic (P1), early symptomatic (P7), and late-symptomatic (P13). Compared to littermate control mice, SMA mice showed a time-dependent increase in the number of exons showing differential expression, with minimal differences between genotypes at P1 and P7, but substantial variation in late-symptomatic (P13) mice. Gene ontology analysis revealed differences in pathways associated with neuronal development as well as cellular injury. Validation of selected targets by RT-PCR confirmed the array findings and was in keeping with a shift between physiologically occurring mRNA isoforms. We conclude that the majority of splicing changes occur late in SMA and may represent a secondary effect of cell injury, though we cannot rule out significant early changes in a small number of transcripts crucial to motor neuron survival.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model.Nature. 2011 Oct 5;478(7367):123-6. doi: 10.1038/nature10485. Nature. 2011. PMID: 21979052 Free PMC article.
-
SMN deficiency alters Nrxn2 expression and splicing in zebrafish and mouse models of spinal muscular atrophy.Hum Mol Genet. 2014 Apr 1;23(7):1754-70. doi: 10.1093/hmg/ddt567. Epub 2013 Nov 11. Hum Mol Genet. 2014. PMID: 24218366
-
Nusinersen ameliorates motor function and prevents motoneuron Cajal body disassembly and abnormal poly(A) RNA distribution in a SMA mouse model.Sci Rep. 2020 Jul 1;10(1):10738. doi: 10.1038/s41598-020-67569-3. Sci Rep. 2020. PMID: 32612161 Free PMC article.
-
The contribution of mouse models to understanding the pathogenesis of spinal muscular atrophy.Dis Model Mech. 2011 Jul;4(4):457-67. doi: 10.1242/dmm.007245. Dis Model Mech. 2011. PMID: 21708901 Free PMC article. Review.
-
Therapeutics development for spinal muscular atrophy.NeuroRx. 2006 Apr;3(2):235-45. doi: 10.1016/j.nurx.2006.01.010. NeuroRx. 2006. PMID: 16554261 Free PMC article. Review.
Cited by
-
Antisense oligonucleotides for the treatment of spinal muscular atrophy.Hum Gene Ther. 2013 May;24(5):489-98. doi: 10.1089/hum.2012.225. Hum Gene Ther. 2013. PMID: 23544870 Free PMC article. Review.
-
SMN - A chaperone for nuclear RNP social occasions?RNA Biol. 2017 Jun 3;14(6):701-711. doi: 10.1080/15476286.2016.1236168. Epub 2016 Sep 20. RNA Biol. 2017. PMID: 27648855 Free PMC article. Review.
-
TSUNAMI: an antisense method to phenocopy splicing-associated diseases in animals.Genes Dev. 2012 Aug 15;26(16):1874-84. doi: 10.1101/gad.197418.112. Genes Dev. 2012. PMID: 22895255 Free PMC article.
-
Single-cell RNA sequencing reveals dysregulation of spinal cord cell types in a severe spinal muscular atrophy mouse model.PLoS Genet. 2022 Sep 8;18(9):e1010392. doi: 10.1371/journal.pgen.1010392. eCollection 2022 Sep. PLoS Genet. 2022. PMID: 36074806 Free PMC article.
-
Mutations in the survival motor neuron (SMN) protein alter the dynamic nature of nuclear bodies.Neuromolecular Med. 2011 Mar;13(1):77-87. doi: 10.1007/s12017-010-8139-1. Epub 2010 Nov 17. Neuromolecular Med. 2011. PMID: 21082361
References
-
- Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA). Hum Mutat. 2000;15:228–237. - PubMed
-
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. - PubMed
-
- Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16:265–269. - PubMed
-
- Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8:1177–1183. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

