A novel type of E3 ligase for the Ufm1 conjugation system
- PMID: 20018847
- PMCID: PMC2820770
- DOI: 10.1074/jbc.M109.036814
A novel type of E3 ligase for the Ufm1 conjugation system
Abstract
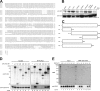
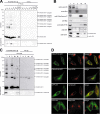
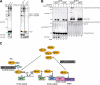
The ubiquitin fold modifier 1 (Ufm1) is the most recently discovered ubiquitin-like modifier whose conjugation (ufmylation) system is conserved in multicellular organisms. Ufm1 is known to covalently attach with cellular protein(s) via a specific E1-activating enzyme (Uba5) and an E2-conjugating enzyme (Ufc1), but its E3-ligating enzyme(s) as well as the target protein(s) remain unknown. Herein, we report both a novel E3 ligase for Ufm1, designated Ufl1, and an Ufm1-specific substrate ligated by Ufl1, C20orf116. Ufm1 was covalently conjugated with C20orf116. Although Ufl1 has no obvious sequence homology to any other known E3s for ubiquitin and ubiquitin-like modifiers, the C20orf116 x Ufm1 formation was greatly accelerated by Ufl1. The C20orf116 x Ufm1 conjugate was cleaved by Ufm1-specific proteases, implying the reversibility of ufmylation. The conjugation was abundant in the liver and lungs of Ufm1-transgenic mice, fractionated into membrane fraction, and impaired in Uba5 knock-out cells. Intriguingly, immunological analysis revealed localizations of Ufl1 and C20orf116 mainly to the endoplasmic reticulum. Our results provide novel insights into the Ufm1 system involved in cellular regulation of multicellular organisms.
Figures



Similar articles
-
UFMylation: A Unique & Fashionable Modification for Life.Genomics Proteomics Bioinformatics. 2016 Jun;14(3):140-146. doi: 10.1016/j.gpb.2016.04.001. Epub 2016 May 20. Genomics Proteomics Bioinformatics. 2016. PMID: 27212118 Free PMC article. Review.
-
Deficiency of Murine UFM1-Specific E3 Ligase Causes Microcephaly and Inflammation.Mol Neurobiol. 2022 Oct;59(10):6363-6372. doi: 10.1007/s12035-022-02979-0. Epub 2022 Aug 6. Mol Neurobiol. 2022. PMID: 35931931
-
UFL1, a UFMylation E3 ligase, plays a crucial role in multiple cellular stress responses.Front Endocrinol (Lausanne). 2023 Feb 10;14:1123124. doi: 10.3389/fendo.2023.1123124. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 36843575 Free PMC article. Review.
-
Optimized protocol to detect protein UFMylation in cells and in vitro via immunoblotting.STAR Protoc. 2022 Jan 6;3(1):101074. doi: 10.1016/j.xpro.2021.101074. eCollection 2022 Mar 18. STAR Protoc. 2022. PMID: 35036955 Free PMC article.
-
Ufl1/RCAD, a Ufm1 E3 ligase, has an intricate connection with ER stress.Int J Biol Macromol. 2019 Aug 15;135:760-767. doi: 10.1016/j.ijbiomac.2019.05.170. Epub 2019 May 23. Int J Biol Macromol. 2019. PMID: 31129212 Review.
Cited by
-
Tackling Drug Resistance in EGFR Exon 20 Insertion Mutant Lung Cancer.Pharmgenomics Pers Med. 2021 Mar 9;14:301-317. doi: 10.2147/PGPM.S242045. eCollection 2021. Pharmgenomics Pers Med. 2021. PMID: 33727854 Free PMC article. Review.
-
Ufl1 deficiency causes skin pigmentation by up-regulation of Endothelin-1.Front Cell Dev Biol. 2022 Sep 2;10:961675. doi: 10.3389/fcell.2022.961675. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36120581 Free PMC article.
-
Deletion of mitochondrial associated ubiquitin fold modifier protein Ufm1 in Leishmania donovani results in loss of β-oxidation of fatty acids and blocks cell division in the amastigote stage.Mol Microbiol. 2012 Oct;86(1):187-98. doi: 10.1111/j.1365-2958.2012.08183.x. Epub 2012 Aug 16. Mol Microbiol. 2012. PMID: 22897198 Free PMC article.
-
Structural study of UFL1-UFC1 interaction uncovers the role of UFL1 N-terminal helix in ufmylation.EMBO Rep. 2023 Dec 6;24(12):e56920. doi: 10.15252/embr.202356920. Epub 2023 Nov 21. EMBO Rep. 2023. PMID: 37988244 Free PMC article.
-
Ufm1-Specific Ligase Ufl1 Regulates Endoplasmic Reticulum Homeostasis and Protects Against Heart Failure.Circ Heart Fail. 2018 Oct;11(10):e004917. doi: 10.1161/CIRCHEARTFAILURE.118.004917. Circ Heart Fail. 2018. PMID: 30354401 Free PMC article.
References
-
- Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 - PubMed
-
- Mukhopadhyay D., Riezman H. (2007) Science 315, 201–205 - PubMed
-
- Pickart C. M., Eddins M. J. (2004) Biochim. Biophys. Acta 1695, 55–72 - PubMed
-
- Groettrup M., Pelzer C., Schmidtke G., Hofmann K. (2008) Trends Biochem. Sci. 33, 230–237 - PubMed
-
- Jin J., Li X., Gygi S. P., Harper J. W. (2007) Nature 447, 1135–1138 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

