Functional screening identifies CRLF2 in precursor B-cell acute lymphoblastic leukemia
- PMID: 20018760
- PMCID: PMC2806782
- DOI: 10.1073/pnas.0911726107
Functional screening identifies CRLF2 in precursor B-cell acute lymphoblastic leukemia
Abstract
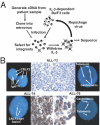
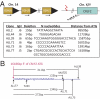
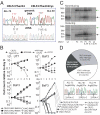
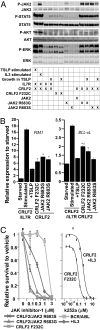
The prognosis for adults with precursor B-cell acute lymphoblastic leukemia (B-ALL) remains poor, in part from a lack of therapeutic targets. We identified the type I cytokine receptor subunit CRLF2 in a functional screen for B-ALL-derived mRNA transcripts that can substitute for IL3 signaling. We demonstrate that CRLF2 is overexpressed in approximately 15% of adult and high-risk pediatric B-ALL that lack MLL, TCF3, TEL, and BCR/ABL rearrangements, but not in B-ALL with these rearrangements or other lymphoid malignancies. CRLF2 overexpression can result from translocation with the IGH locus or intrachromosomal deletion and is associated with poor outcome. CRLF2 overexpressing B-ALLs share a transcriptional signature that significantly overlaps with a BCR/ABL signature, and is enriched for genes involved in cytokine receptor and JAK-STAT signaling. In a subset of cases, CRLF2 harbors a Phe232Cys gain-of-function mutation that promotes constitutive dimerization and cytokine independent growth. A mutually exclusive subset harbors activating mutations in JAK2. In fact, all 22 B-ALLs with mutant JAK2 that we analyzed overexpress CRLF2, distinguishing CRLF2 as the key scaffold for mutant JAK2 signaling in B-ALL. Expression of WT CRLF2 with mutant JAK2 also promotes cytokine independent growth that, unlike CRLF2 Phe232Cys or ligand-induced signaling by WT CRLF2, is accompanied by JAK2 phosphorylation. Finally, cells dependent on CRLF2 signaling are sensitive to small molecule inhibitors of either JAKs or protein kinase C family kinases. Together, these findings implicate CRLF2 as an important factor in B-ALL with diagnostic, prognostic, and therapeutic implications.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
CRLF2 and JAK2 in B-progenitor acute lymphoblastic leukemia: a novel association in oncogenesis.Cancer Res. 2010 Oct 1;70(19):7347-52. doi: 10.1158/0008-5472.CAN-10-1528. Epub 2010 Aug 31. Cancer Res. 2010. PMID: 20807819 Free PMC article. Review.
-
Differences in signaling through the B-cell leukemia oncoprotein CRLF2 in response to TSLP and through mutant JAK2.Blood. 2012 Oct 4;120(14):2853-63. doi: 10.1182/blood-2012-02-413252. Epub 2012 Aug 20. Blood. 2012. PMID: 22915648 Free PMC article.
-
CRLF2-Positive B-Cell Acute Lymphoblastic Leukemia in Adult Patients: A Single-Institution Experience.Am J Clin Pathol. 2017 Apr 1;147(4):357-363. doi: 10.1093/ajcp/aqx005. Am J Clin Pathol. 2017. PMID: 28340183
-
Prognostic significance of CRLF2 overexpression and JAK2 mutation in Egyptian pediatric patients with B-precursor acute lymphoblastic leukemia.Clin Lymphoma Myeloma Leuk. 2022 Jun;22(6):e376-e385. doi: 10.1016/j.clml.2021.12.006. Epub 2021 Dec 11. Clin Lymphoma Myeloma Leuk. 2022. PMID: 34987014
-
Understanding the biology of CRLF2-overexpressing acute lymphoblastic leukemia.Crit Rev Oncog. 2011;16(1-2):13-24. doi: 10.1615/critrevoncog.v16.i1-2.30. Crit Rev Oncog. 2011. PMID: 22150304 Free PMC article. Review.
Cited by
-
BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia.Blood. 2012 Oct 4;120(14):2843-52. doi: 10.1182/blood-2012-02-413021. Epub 2012 Aug 17. Blood. 2012. PMID: 22904298 Free PMC article.
-
Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer.Genome Res. 2012 Nov;22(11):2120-9. doi: 10.1101/gr.137596.112. Epub 2012 Oct 1. Genome Res. 2012. PMID: 23028188 Free PMC article.
-
CRLF2 and JAK2 in B-progenitor acute lymphoblastic leukemia: a novel association in oncogenesis.Cancer Res. 2010 Oct 1;70(19):7347-52. doi: 10.1158/0008-5472.CAN-10-1528. Epub 2010 Aug 31. Cancer Res. 2010. PMID: 20807819 Free PMC article. Review.
-
JAK2 Alterations in Acute Lymphoblastic Leukemia: Molecular Insights for Superior Precision Medicine Strategies.Front Cell Dev Biol. 2022 Jul 12;10:942053. doi: 10.3389/fcell.2022.942053. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35903543 Free PMC article. Review.
-
Constitutive Ras signaling and Ink4a/Arf inactivation cooperate during the development of B-ALL in mice.Blood Adv. 2017 Nov 21;1(25):2361-2374. doi: 10.1182/bloodadvances.2017012211. eCollection 2017 Nov 28. Blood Adv. 2017. PMID: 29296886 Free PMC article.
References
-
- Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. - PubMed
-
- Mullighan CG, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. - PubMed
-
- Chiaretti S, et al. Gene expression profiles of B-lineage adult acute lymphocytic leukemia reveal genetic patterns that identify lineage derivation and distinct mechanisms of transformation. Clin Cancer Res. 2005;11:7209–7219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

