Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks
- PMID: 20016603
- PMCID: PMC2904806
- DOI: 10.1038/nature08657
Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks
Abstract
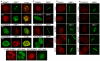
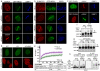
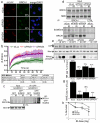
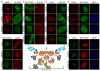
DNA double-strand breaks (DSBs) are highly cytotoxic lesions that are generated by ionizing radiation and various DNA-damaging chemicals. Following DSB formation, cells activate the DNA-damage response (DDR) protein kinases ATM, ATR and DNA-PK (also known as PRKDC). These then trigger histone H2AX (also known as H2AFX) phosphorylation and the accumulation of proteins such as MDC1, 53BP1 (also known as TP53BP1), BRCA1, CtIP (also known as RBBP8), RNF8 and RNF168/RIDDLIN into ionizing radiation-induced foci (IRIF) that amplify DSB signalling and promote DSB repair. Attachment of small ubiquitin-related modifier (SUMO) to target proteins controls diverse cellular functions. Here, we show that SUMO1, SUMO2 and SUMO3 accumulate at DSB sites in mammalian cells, with SUMO1 and SUMO2/3 accrual requiring the E3 ligase enzymes PIAS4 and PIAS1. We also establish that PIAS1 and PIAS4 are recruited to damage sites via mechanisms requiring their SAP domains, and are needed for the productive association of 53BP1, BRCA1 and RNF168 with such regions. Furthermore, we show that PIAS1 and PIAS4 promote DSB repair and confer ionizing radiation resistance. Finally, we establish that PIAS1 and PIAS4 are required for effective ubiquitin-adduct formation mediated by RNF8, RNF168 and BRCA1 at sites of DNA damage. These findings thus identify PIAS1 and PIAS4 as components of the DDR and reveal how protein recruitment to DSB sites is controlled by coordinated SUMOylation and ubiquitylation.
Figures
Comment in
-
DNA repair: A heavyweight joins the fray.Nature. 2009 Dec 17;462(7275):857-8. doi: 10.1038/462857a. Nature. 2009. PMID: 20016586 No abstract available.
Similar articles
-
DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger.J Cell Biol. 2012 Apr 16;197(2):179-87. doi: 10.1083/jcb.201106152. J Cell Biol. 2012. PMID: 22508508 Free PMC article.
-
Roles of the SUMO-related enzymes, PIAS1, PIAS4, and RNF4, in DNA double-strand break repair by homologous recombination.Biochem Biophys Res Commun. 2022 Feb 5;591:95-101. doi: 10.1016/j.bbrc.2021.12.099. Epub 2021 Dec 30. Biochem Biophys Res Commun. 2022. PMID: 35007836
-
RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair.Genes Dev. 2012 Jun 1;26(11):1179-95. doi: 10.1101/gad.188284.112. Genes Dev. 2012. PMID: 22661229 Free PMC article.
-
The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks.FEBS Lett. 2011 Sep 16;585(18):2914-9. doi: 10.1016/j.febslet.2011.05.056. Epub 2011 Jun 12. FEBS Lett. 2011. PMID: 21664912 Review.
-
Opposing roles of RNF8/RNF168 and deubiquitinating enzymes in ubiquitination-dependent DNA double-strand break response signaling and DNA-repair pathway choice.J Radiat Res. 2016 Aug;57 Suppl 1(Suppl 1):i33-i40. doi: 10.1093/jrr/rrw027. Epub 2016 Mar 16. J Radiat Res. 2016. PMID: 26983989 Free PMC article. Review.
Cited by
-
PML Degradation: Multiple Ways to Eliminate PML.Front Oncol. 2013 Mar 22;3:60. doi: 10.3389/fonc.2013.00060. eCollection 2013. Front Oncol. 2013. PMID: 23526763 Free PMC article.
-
System-wide Analysis of SUMOylation Dynamics in Response to Replication Stress Reveals Novel Small Ubiquitin-like Modified Target Proteins and Acceptor Lysines Relevant for Genome Stability.Mol Cell Proteomics. 2015 May;14(5):1419-34. doi: 10.1074/mcp.O114.044792. Epub 2015 Mar 9. Mol Cell Proteomics. 2015. PMID: 25755297 Free PMC article.
-
Adenovirus regulates sumoylation of Mre11-Rad50-Nbs1 components through a paralog-specific mechanism.J Virol. 2012 Sep;86(18):9656-65. doi: 10.1128/JVI.01273-12. Epub 2012 Jun 27. J Virol. 2012. PMID: 22740413 Free PMC article.
-
DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger.J Cell Biol. 2012 Apr 16;197(2):179-87. doi: 10.1083/jcb.201106152. J Cell Biol. 2012. PMID: 22508508 Free PMC article.
-
SUMO2/3 modification of cyclin E contributes to the control of replication origin firing.Nat Commun. 2013;4:1850. doi: 10.1038/ncomms2875. Nat Commun. 2013. PMID: 23673635 Free PMC article.
References
-
- Stucki M, Jackson SP. MDC1/NFBD1: a key regulator of the DNA damage response in higher eukaryotes. DNA Repair (Amst) 2004;3:953–957. - PubMed
-
- Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–958. - PubMed
-
- Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. - PubMed
-
- Meulmeester E, Melchior F. Cell biology: SUMO. Nature. 2008;452:709–711. - PubMed
-
- Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

