The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants
- PMID: 20015970
- PMCID: PMC2847217
- DOI: 10.1093/nar/gkp1137
The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants
Abstract
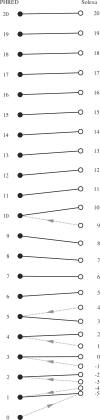
FASTQ has emerged as a common file format for sharing sequencing read data combining both the sequence and an associated per base quality score, despite lacking any formal definition to date, and existing in at least three incompatible variants. This article defines the FASTQ format, covering the original Sanger standard, the Solexa/Illumina variants and conversion between them, based on publicly available information such as the MAQ documentation and conventions recently agreed by the Open Bioinformatics Foundation projects Biopython, BioPerl, BioRuby, BioJava and EMBOSS. Being an open access publication, it is hoped that this description, with the example files provided as Supplementary Data, will serve in future as a reference for this important file format.
Figures
Similar articles
-
ClinQC: a tool for quality control and cleaning of Sanger and NGS data in clinical research.BMC Bioinformatics. 2016 Feb 2;17:56. doi: 10.1186/s12859-016-0915-y. BMC Bioinformatics. 2016. PMID: 26830926 Free PMC article.
-
XS: a FASTQ read simulator.BMC Res Notes. 2014 Jan 16;7:40. doi: 10.1186/1756-0500-7-40. BMC Res Notes. 2014. PMID: 24433564 Free PMC article.
-
Sharing Programming Resources Between Bio* Projects.Methods Mol Biol. 2019;1910:747-766. doi: 10.1007/978-1-4939-9074-0_25. Methods Mol Biol. 2019. PMID: 31278684 Free PMC article.
-
Introduction to bioinformatics.Mol Nutr Food Res. 2006 Jul;50(7):610-9. doi: 10.1002/mnfr.200500273. Mol Nutr Food Res. 2006. PMID: 16810733 Review.
-
A brief history of bioinformatics.Brief Bioinform. 2019 Nov 27;20(6):1981-1996. doi: 10.1093/bib/bby063. Brief Bioinform. 2019. PMID: 30084940 Review.
Cited by
-
Modeling methyl-sensitive transcription factor motifs with an expanded epigenetic alphabet.Genome Biol. 2024 Jan 8;25(1):11. doi: 10.1186/s13059-023-03070-0. Genome Biol. 2024. PMID: 38191487 Free PMC article.
-
Fitness analyses of all possible point mutations for regions of genes in yeast.Nat Protoc. 2012 Jun 21;7(7):1382-96. doi: 10.1038/nprot.2012.069. Nat Protoc. 2012. PMID: 22722372 Free PMC article.
-
Phylogenomics, taxonomy and morphological characters of the Microdochiaceae (Xylariales, Sordariomycetes).MycoKeys. 2024 Jul 3;106:303-325. doi: 10.3897/mycokeys.106.127355. eCollection 2024. MycoKeys. 2024. PMID: 38993357 Free PMC article.
-
MaAzaR, a Zn2Cys6/Fungus-Specific Transcriptional Factor, Is Involved in Stress Tolerance and Conidiation Pattern Shift in Metarhizium acridum.J Fungi (Basel). 2024 Jul 4;10(7):468. doi: 10.3390/jof10070468. J Fungi (Basel). 2024. PMID: 39057353 Free PMC article.
-
Parallel evolutionary dynamics of adaptive diversification in Escherichia coli.PLoS Biol. 2013;11(2):e1001490. doi: 10.1371/journal.pbio.1001490. Epub 2013 Feb 19. PLoS Biol. 2013. PMID: 23431270 Free PMC article.
References
-
- Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. - PubMed
-
- Pandey V, Nutter RC, E EP. Applied Biosystems SOLiD system: ligation-based sequencing. In: Janitz M, editor. Next Generation Genome Sequencing: Towards Personalized Medicine. Wiley; 2008. pp. 29–41.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

