FOG-1-mediated recruitment of NuRD is required for cell lineage re-enforcement during haematopoiesis
- PMID: 20010697
- PMCID: PMC2824465
- DOI: 10.1038/emboj.2009.368
FOG-1-mediated recruitment of NuRD is required for cell lineage re-enforcement during haematopoiesis
Abstract
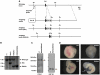
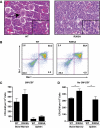
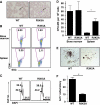
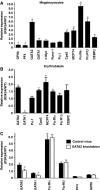
The transcriptional co-factor Friend of GATA1 (FOG-1) has been shown to interact with subunits of the nucleosome remodelling and histone deacetylase (NuRD) complex through a specific motif located at its N-terminus. To test the importance of FOG-1/NuRD interaction for haematopoiesis in vivo, we generated mice with a mutation that specifically disrupts FOG-1/NuRD interaction (FOG-1(R3K5A)). Homozygous FOG-1(R3K5A) mice were found to have splenomegaly, extramedullary erythropoiesis, granulocytosis and thrombocytopaenia secondary to a block in megakaryocyte maturation. FOG-1(R3K5A/R3K5A) megakaryocytes and erythroid progenitors expressed increased levels of GATA2, showing that FOG-1/NuRD interaction is required for the earlier described 'GATA Switch'. In addition, ablation of FOG-1/NuRD interaction led to inappropriate expression of mast cell and eosinophil-specific genes in the megakaryocyte and erythroid lineages. Chromatin immunoprecipitation experiments revealed that the NuRD complex was not properly recruited to a mast cell gene promoter in FOG-1(R3K5A/R3K5A) megakaryocytes, suggesting that FOG-1/NuRD interaction is required for the direct suppression of mast cell gene expression. Taken together, these results underscore the importance of the FOG-1/NuRD interaction for the re-enforcement of lineage commitment during erythropoiesis and megakaryopoiesis in vivo.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
FOG1 requires NuRD to promote hematopoiesis and maintain lineage fidelity within the megakaryocytic-erythroid compartment.Blood. 2010 Mar 18;115(11):2156-66. doi: 10.1182/blood-2009-10-251280. Epub 2010 Jan 11. Blood. 2010. PMID: 20065294 Free PMC article.
-
NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development.EMBO J. 2010 Jan 20;29(2):442-56. doi: 10.1038/emboj.2009.336. Epub 2009 Nov 19. EMBO J. 2010. PMID: 19927129 Free PMC article.
-
FOG-2 mediated recruitment of the NuRD complex regulates cardiomyocyte proliferation during heart development.Dev Biol. 2014 Nov 1;395(1):50-61. doi: 10.1016/j.ydbio.2014.08.030. Epub 2014 Sep 6. Dev Biol. 2014. PMID: 25196150 Free PMC article.
-
Mi-2/NuRD chromatin remodeling complexes regulate B and T-lymphocyte development and function.Immunol Rev. 2014 Sep;261(1):126-40. doi: 10.1111/imr.12209. Immunol Rev. 2014. PMID: 25123281 Free PMC article. Review.
-
The Mi-2/NuRD complex: a critical epigenetic regulator of hematopoietic development, differentiation and cancer.Epigenetics. 2009 Nov 16;4(8):532-6. doi: 10.4161/epi.4.8.10108. Epub 2009 Nov 16. Epigenetics. 2009. PMID: 19923891 Review.
Cited by
-
Signal transduction pathways, intrinsic regulators, and the control of cell fate choice.Biochim Biophys Acta. 2013 Feb;1830(2):2375-84. doi: 10.1016/j.bbagen.2012.06.005. Epub 2012 Jun 15. Biochim Biophys Acta. 2013. PMID: 22705942 Free PMC article. Review.
-
A Genomic Link From Heart Failure to Atrial Fibrillation Risk: FOG2 Modulates a TBX5/GATA4-Dependent Atrial Gene Regulatory Network.Circulation. 2024 Apr 9;149(15):1205-1230. doi: 10.1161/CIRCULATIONAHA.123.066804. Epub 2024 Jan 8. Circulation. 2024. PMID: 38189150
-
Overlapping functions of Hdac1 and Hdac2 in cell cycle regulation and haematopoiesis.EMBO J. 2010 Aug 4;29(15):2586-97. doi: 10.1038/emboj.2010.136. Epub 2010 Jun 22. EMBO J. 2010. PMID: 20571512 Free PMC article.
-
Hedgehog signaling from the Posterior Signaling Center maintains U-shaped expression and a prohemocyte population in Drosophila.Dev Biol. 2018 Sep 1;441(1):132-145. doi: 10.1016/j.ydbio.2018.06.020. Epub 2018 Jul 11. Dev Biol. 2018. PMID: 29966604 Free PMC article.
-
PICKLE is a CHD subfamily II ATP-dependent chromatin remodeling factor.Biochim Biophys Acta. 2013 Feb;1829(2):199-210. doi: 10.1016/j.bbagrm.2012.10.011. Epub 2012 Nov 2. Biochim Biophys Acta. 2013. PMID: 23128324 Free PMC article.
References
-
- Cismasiu VB, Adamo K, Gecewicz J, Duque J, Lin Q, Avram D (2005) BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene 24: 6753–6764 - PubMed
-
- Crispino JD, Lodish MB, MacKay JP, Orkin SH (1999) Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol Cell 3: 219–228 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

