Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease
- PMID: 20008565
- PMCID: PMC2806317
- DOI: 10.1083/jcb.200908115
Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease
Abstract
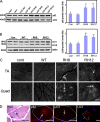
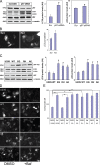
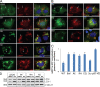
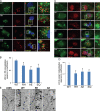
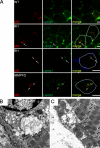
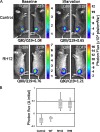
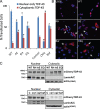
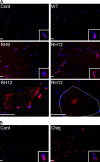
Mutations in valosin-containing protein (VCP) cause inclusion body myopathy (IBM), Paget's disease of the bone, and frontotemporal dementia (IBMPFD). Patient muscle has degenerating fibers, rimmed vacuoles (RVs), and sarcoplasmic inclusions containing ubiquitin and TDP-43 (TARDNA-binding protein 43). In this study, we find that IBMPFD muscle also accumulates autophagosome-associated proteins, Map1-LC3 (LC3), and p62/sequestosome, which localize to RVs. To test whether VCP participates in autophagy, we silenced VCP or expressed adenosine triphosphatase-inactive VCP. Under basal conditions, loss of VCP activity results in autophagosome accumulation. After autophagic induction, these autophagosomes fail to mature into autolysosomes and degrade LC3. Similarly, IBMPFD mutant VCP expression in cells and animals leads to the accumulation of nondegradative autophagosomes that coalesce at RVs and fail to degrade aggregated proteins. Interestingly, TDP-43 accumulates in the cytosol upon autophagic inhibition, similar to that seen after IBMPFD mutant expression. These data implicate VCP in autophagy and suggest that impaired autophagy explains the pathology seen in IBMPFD muscle, including TDP-43 accumulation.
Figures

Similar articles
-
VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD.Autophagy. 2010 Feb;6(2):217-27. doi: 10.4161/auto.6.2.11014. Epub 2010 Feb 22. Autophagy. 2010. PMID: 20104022 Free PMC article.
-
The multiple faces of valosin-containing protein-associated diseases: inclusion body myopathy with Paget's disease of bone, frontotemporal dementia, and amyotrophic lateral sclerosis.J Mol Neurosci. 2011 Nov;45(3):522-31. doi: 10.1007/s12031-011-9627-y. Epub 2011 Sep 3. J Mol Neurosci. 2011. PMID: 21892620 Review.
-
Transgenic mice expressing mutant forms VCP/p97 recapitulate the full spectrum of IBMPFD including degeneration in muscle, brain and bone.Hum Mol Genet. 2010 May 1;19(9):1741-55. doi: 10.1093/hmg/ddq050. Epub 2010 Feb 10. Hum Mol Genet. 2010. PMID: 20147319
-
Neuronal-specific overexpression of a mutant valosin-containing protein associated with IBMPFD promotes aberrant ubiquitin and TDP-43 accumulation and cognitive dysfunction in transgenic mice.Am J Pathol. 2013 Aug;183(2):504-15. doi: 10.1016/j.ajpath.2013.04.014. Epub 2013 Jun 5. Am J Pathol. 2013. PMID: 23747512 Free PMC article.
-
Valosin-containing protein disease: inclusion body myopathy with Paget's disease of the bone and fronto-temporal dementia.Neuromuscul Disord. 2009 May;19(5):308-15. doi: 10.1016/j.nmd.2009.01.009. Epub 2009 Apr 19. Neuromuscul Disord. 2009. PMID: 19380227 Free PMC article. Review.
Cited by
-
New functions of a known autophagy regulator: VCP and autophagy initiation.Autophagy. 2021 May;17(5):1063-1064. doi: 10.1080/15548627.2021.1905974. Epub 2021 Mar 31. Autophagy. 2021. PMID: 33784947 Free PMC article.
-
Structural and functional deviations in disease-associated p97 mutants.J Struct Biol. 2012 Aug;179(2):83-92. doi: 10.1016/j.jsb.2012.04.024. Epub 2012 May 8. J Struct Biol. 2012. PMID: 22579784 Free PMC article. Review.
-
AAA ATPase p97/VCP is essential for TRIM21-mediated virus neutralization.Proc Natl Acad Sci U S A. 2012 Nov 27;109(48):19733-8. doi: 10.1073/pnas.1210659109. Epub 2012 Oct 22. Proc Natl Acad Sci U S A. 2012. PMID: 23091005 Free PMC article.
-
A Fine Balance of Dietary Lipids Improves Pathology of a Murine Model of VCP-Associated Multisystem Proteinopathy.PLoS One. 2015 Jul 2;10(7):e0131995. doi: 10.1371/journal.pone.0131995. eCollection 2015. PLoS One. 2015. PMID: 26134519 Free PMC article.
-
Altered cofactor regulation with disease-associated p97/VCP mutations.Proc Natl Acad Sci U S A. 2015 Apr 7;112(14):E1705-14. doi: 10.1073/pnas.1418820112. Epub 2015 Mar 16. Proc Natl Acad Sci U S A. 2015. PMID: 25775548 Free PMC article.
References
-
- Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E.M., Isaacs A., Brech A., Stenmark H., Simonsen A. 2007. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179:485–500 10.1083/jcb.200702115 - DOI - PMC - PubMed
-
- Forman M.S., Mackenzie I.R., Cairns N.J., Swanson E., Boyer P.J., Drachman D.A., Jhaveri B.S., Karlawish J.H., Pestronk A., Smith T.W., et al. 2006. Novel ubiquitin neuropathology in frontotemporal dementia with valosin-containing protein gene mutations. J. Neuropathol. Exp. Neurol. 65:571–581 10.1097/00005072-200606000-00005 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

