Nucleolin associates with the human cytomegalovirus DNA polymerase accessory subunit UL44 and is necessary for efficient viral replication
- PMID: 20007282
- PMCID: PMC2812382
- DOI: 10.1128/JVI.01510-09
Nucleolin associates with the human cytomegalovirus DNA polymerase accessory subunit UL44 and is necessary for efficient viral replication
Abstract
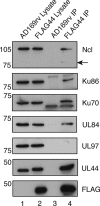
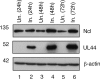
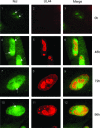
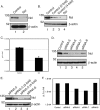
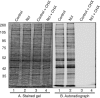
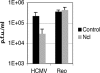
In the eukaryotic cell, DNA replication entails the interaction of multiple proteins with the DNA polymerase processivity factor PCNA. As the structure of the presumptive human cytomegalovirus (HCMV) DNA polymerase processivity factor UL44 is highly homologous to that of PCNA, we hypothesized that UL44 also interacts with numerous proteins. To investigate this possibility, recombinant HCMV expressing FLAG-tagged UL44 was generated and used to immunoprecipitate UL44 and associated proteins from infected cell lysates. Unexpectedly, nucleolin, a major protein component of the nucleolus, was identified among these proteins by mass spectrometry and Western blotting. The association of nucleolin and UL44 in infected cell lysate was confirmed by reciprocal coimmunoprecipitation in the presence and absence of nuclease. Western blotting and immunofluorescence assays demonstrated that the level of nucleolin increases during infection and that nucleolin becomes distributed throughout the nucleus. Furthermore, the colocalization of nucleolin and UL44 in infected cell nuclei was observed by immunofluorescence assays. Assays of HCMV-infected cells treated with small interfering RNA (siRNA) targeting nucleolin mRNA indicated that nucleolin was required for efficient virus production, viral DNA synthesis, and the expression of a late viral protein, with a correlation between the efficacy of knockdown and the effect on virus replication. In contrast, the level of neither global protein synthesis nor the replication of an unrelated virus (reovirus) was reduced in siRNA-treated cells. Taken together, our results indicate an association of nucleolin and UL44 in HCMV-infected cells and a role for nucleolin in viral DNA synthesis.
Figures



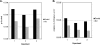
Similar articles
-
Host cell nucleolin is required to maintain the architecture of human cytomegalovirus replication compartments.mBio. 2012 Feb 7;3(1):e00301-11. doi: 10.1128/mBio.00301-11. Print 2012. mBio. 2012. PMID: 22318319 Free PMC article.
-
Dynamic and nucleolin-dependent localization of human cytomegalovirus UL84 to the periphery of viral replication compartments and nucleoli.J Virol. 2014 Oct;88(20):11738-47. doi: 10.1128/JVI.01889-14. Epub 2014 Jul 30. J Virol. 2014. PMID: 25078694 Free PMC article.
-
Analysis of the association of the human cytomegalovirus DNA polymerase subunit UL44 with the viral DNA replication factor UL84.J Virol. 2009 Aug;83(15):7581-9. doi: 10.1128/JVI.00663-09. Epub 2009 May 20. J Virol. 2009. PMID: 19457994 Free PMC article.
-
A mutation deleting sequences encoding the amino terminus of human cytomegalovirus UL84 impairs interaction with UL44 and capsid localization.J Virol. 2012 Oct;86(20):11066-77. doi: 10.1128/JVI.01379-12. Epub 2012 Aug 1. J Virol. 2012. PMID: 22855486 Free PMC article.
-
Nuts and bolts of human cytomegalovirus lytic DNA replication.Curr Top Microbiol Immunol. 2008;325:153-66. doi: 10.1007/978-3-540-77349-8_9. Curr Top Microbiol Immunol. 2008. PMID: 18637505 Review.
Cited by
-
Nucleolin is important for Epstein-Barr virus nuclear antigen 1-mediated episome binding, maintenance, and transcription.Proc Natl Acad Sci U S A. 2014 Jan 7;111(1):243-8. doi: 10.1073/pnas.1321800111. Epub 2013 Dec 16. Proc Natl Acad Sci U S A. 2014. PMID: 24344309 Free PMC article.
-
Characterisation of a human antibody that potentially links cytomegalovirus infection with systemic lupus erythematosus.Sci Rep. 2019 Jul 10;9(1):9998. doi: 10.1038/s41598-019-46329-y. Sci Rep. 2019. PMID: 31292492 Free PMC article.
-
Nucleolin interacts with the feline calicivirus 3' untranslated region and the protease-polymerase NS6 and NS7 proteins, playing a role in virus replication.J Virol. 2011 Aug;85(16):8056-68. doi: 10.1128/JVI.01878-10. Epub 2011 Jun 15. J Virol. 2011. PMID: 21680514 Free PMC article.
-
Association of human cytomegalovirus proteins IRS1 and TRS1 with the viral DNA polymerase accessory subunit UL44.J Gen Virol. 2010 Sep;91(Pt 9):2167-75. doi: 10.1099/vir.0.022640-0. Epub 2010 May 5. J Gen Virol. 2010. PMID: 20444996 Free PMC article.
-
Virus-host protein interactions as footprints of human cytomegalovirus replication.Curr Opin Virol. 2022 Feb;52:135-147. doi: 10.1016/j.coviro.2021.11.016. Epub 2021 Dec 16. Curr Opin Virol. 2022. PMID: 34923282 Free PMC article. Review.
References
-
- Appleton, B. A., J. Brooks, A. Loregian, D. J. Filman, D. M. Coen, and J. M. Hogle. 2006. Crystal structure of the cytomegalovirus DNA polymerase subunit UL44 in complex with the C terminus from the catalytic subunit. Differences in structure and function relative to unliganded UL44. J. Biol. Chem. 281:5224-5232. - PubMed
-
- Appleton, B. A., A. Loregian, D. J. Filman, D. M. Coen, and J. M. Hogle. 2004. The cytomegalovirus DNA polymerase subunit UL44 forms a C clamp-shaped dimer. Mol. Cell 15:233-244. - PubMed
-
- Bertrand, L., and A. Pearson. 2008. The conserved N-terminal domain of herpes simplex virus 1 UL24 protein is sufficient to induce the spatial redistribution of nucleolin. J. Gen. Virol. 89:1142-1151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

