Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters
- PMID: 19948768
- PMCID: PMC2802027
- DOI: 10.1261/rna.1873910
Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters
Abstract
Murid herpesvirus 4 (MuHV-4) microRNAs were previously cloned from latently infected tumor cells and predicted to be processed from a series of RNA polymerase III primary transcripts. We detected maturely processed MuHV-4 miRNAs within total RNA from lytically infected cells in vitro and infected tissues ex vivo, using a highly sensitive reverse ligation meditated RT-PCR strategy. We determined that the MuHV-4 microRNAs are biologically active during infection by a luciferase reporter system. We experimentally demonstrated that transcription of the MuHV-4 microRNAs is by RNA polymerase III by alpha-amanitin insensitivity and by specific deletion of the RNA polymerase III type 2-like promoter elements of MuHV-4, resulting in the complete loss of miRNA detection and function. Finally, we demonstrate that these 10 viral miRNAs, each transcribed from highly conserved and novel polymerase III promoter elements, vary markedly in their relative abundance and activity.
Figures
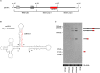


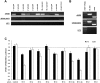


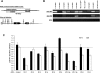

Similar articles
-
A conserved RNA polymerase III promoter required for gammaherpesvirus TMER transcription and microRNA processing.Gene. 2014 Jul 1;544(1):8-18. doi: 10.1016/j.gene.2014.04.026. Epub 2014 Apr 16. Gene. 2014. PMID: 24747015 Free PMC article.
-
A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs.Mol Cell. 2010 Jan 15;37(1):135-42. doi: 10.1016/j.molcel.2009.12.016. Mol Cell. 2010. PMID: 20129062 Free PMC article.
-
Japanese Macaque Rhadinovirus Encodes a Viral MicroRNA Mimic of the miR-17 Family.J Virol. 2016 Sep 29;90(20):9350-63. doi: 10.1128/JVI.01123-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27512057 Free PMC article.
-
Herpesvirus microRNAs: phenotypes and functions.Curr Opin Virol. 2011 Sep;1(3):211-5. doi: 10.1016/j.coviro.2011.04.003. Curr Opin Virol. 2011. PMID: 21927637 Free PMC article. Review.
-
The functions of herpesvirus-encoded microRNAs.Med Microbiol Immunol. 2008 Jun;197(2):261-7. doi: 10.1007/s00430-007-0070-1. Epub 2007 Dec 18. Med Microbiol Immunol. 2008. PMID: 18087721 Free PMC article. Review.
Cited by
-
Viral miRNA regulation of host gene expression.Semin Cell Dev Biol. 2023 Sep 15;146:2-19. doi: 10.1016/j.semcdb.2022.11.007. Epub 2022 Nov 30. Semin Cell Dev Biol. 2023. PMID: 36463091 Free PMC article. Review.
-
DUSP11 activity on triphosphorylated transcripts promotes Argonaute association with noncanonical viral microRNAs and regulates steady-state levels of cellular noncoding RNAs.Genes Dev. 2016 Sep 15;30(18):2076-2092. doi: 10.1101/gad.282616.116. Genes Dev. 2016. PMID: 27798849 Free PMC article.
-
Metazoan MicroRNAs.Cell. 2018 Mar 22;173(1):20-51. doi: 10.1016/j.cell.2018.03.006. Cell. 2018. PMID: 29570994 Free PMC article. Review.
-
Evaluation of immune sensor responses to a viral small noncoding RNA.Front Cell Infect Microbiol. 2024 Oct 8;14:1459256. doi: 10.3389/fcimb.2024.1459256. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39450336 Free PMC article.
-
Diverse virus-host interactions influence RNA-based regulation during γ-herpesvirus infection.Curr Opin Microbiol. 2012 Aug;15(4):506-11. doi: 10.1016/j.mib.2012.05.010. Epub 2012 Jun 9. Curr Opin Microbiol. 2012. PMID: 22682977 Free PMC article. Review.
References
-
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. - PubMed
-
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Blaskovic D, Stanceková M, Svobodová J, Mistríková J. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 1980;24:468. - PubMed
-
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
