X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor
- PMID: 19946266
- PMCID: PMC2861655
- DOI: 10.1038/nature08624
X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor
Abstract
Ionotropic glutamate receptors mediate most excitatory neurotransmission in the central nervous system and function by opening a transmembrane ion channel upon binding of glutamate. Despite their crucial role in neurobiology, the architecture and atomic structure of an intact ionotropic glutamate receptor are unknown. Here we report the crystal structure of the alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA)-sensitive, homotetrameric, rat GluA2 receptor at 3.6 A resolution in complex with a competitive antagonist. The receptor harbours an overall axis of two-fold symmetry with the extracellular domains organized as pairs of local dimers and with the ion channel domain exhibiting four-fold symmetry. A symmetry mismatch between the extracellular and ion channel domains is mediated by two pairs of conformationally distinct subunits, A/C and B/D. Therefore, the stereochemical manner in which the A/C subunits are coupled to the ion channel gate is different from the B/D subunits. Guided by the GluA2 structure and site-directed cysteine mutagenesis, we suggest that GluN1 and GluN2A NMDA (N-methyl-d-aspartate) receptors have a similar architecture, with subunits arranged in a 1-2-1-2 pattern. We exploit the GluA2 structure to develop mechanisms of ion channel activation, desensitization and inhibition by non-competitive antagonists and pore blockers.
Figures
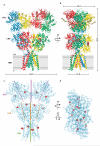

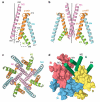
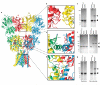


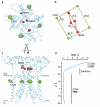
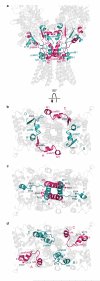
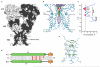
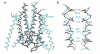
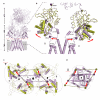
Comment in
-
Neuroscience: Excitatory view of a receptor.Nature. 2009 Dec 10;462(7274):729-31. doi: 10.1038/462729a. Nature. 2009. PMID: 20010675 Free PMC article.
Similar articles
-
Channel opening and gating mechanism in AMPA-subtype glutamate receptors.Nature. 2017 Sep 7;549(7670):60-65. doi: 10.1038/nature23479. Epub 2017 Jul 24. Nature. 2017. PMID: 28737760 Free PMC article.
-
Probing Intersubunit Interfaces in AMPA-subtype Ionotropic Glutamate Receptors.Sci Rep. 2016 Jan 7;6:19082. doi: 10.1038/srep19082. Sci Rep. 2016. PMID: 26739260 Free PMC article.
-
α-Amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) and N-methyl-D-aspartate (NMDA) receptors adopt different subunit arrangements.J Biol Chem. 2013 Jul 26;288(30):21987-98. doi: 10.1074/jbc.M113.469205. Epub 2013 Jun 11. J Biol Chem. 2013. PMID: 23760273 Free PMC article.
-
Structural dynamics in α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor gating.Curr Opin Struct Biol. 2024 Aug;87:102833. doi: 10.1016/j.sbi.2024.102833. Epub 2024 May 10. Curr Opin Struct Biol. 2024. PMID: 38733862 Review.
-
The biochemistry, ultrastructure, and subunit assembly mechanism of AMPA receptors.Mol Neurobiol. 2010 Dec;42(3):161-84. doi: 10.1007/s12035-010-8149-x. Epub 2010 Nov 16. Mol Neurobiol. 2010. PMID: 21080238 Free PMC article. Review.
Cited by
-
α-Lipoic Acid Derivatives as Allosteric Modulators for Targeting AMPA-Type Glutamate Receptors' Gating Modules.Cells. 2022 Nov 15;11(22):3608. doi: 10.3390/cells11223608. Cells. 2022. PMID: 36429036 Free PMC article.
-
Molecular mechanism of parallel fiber-Purkinje cell synapse formation.Front Neural Circuits. 2012 Nov 23;6:90. doi: 10.3389/fncir.2012.00090. eCollection 2012. Front Neural Circuits. 2012. PMID: 23189042 Free PMC article.
-
Correlated conformational dynamics of the human GluN1-GluN2A type N-methyl-D-aspartate (NMDA) receptor.J Mol Model. 2021 May 10;27(6):162. doi: 10.1007/s00894-021-04755-8. J Mol Model. 2021. PMID: 33969428
-
Activation of NMDA receptors and the mechanism of inhibition by ifenprodil.Nature. 2016 Jun 2;534(7605):63-8. doi: 10.1038/nature17679. Epub 2016 May 2. Nature. 2016. PMID: 27135925 Free PMC article.
-
Distinct Subunit Domains Govern Synaptic Stability and Specificity of the Kainate Receptor.Cell Rep. 2016 Jul 12;16(2):531-544. doi: 10.1016/j.celrep.2016.05.093. Epub 2016 Jun 23. Cell Rep. 2016. PMID: 27346345 Free PMC article.
References
-
- Cowan WM, Sudhof TC, Stevens CF, editors. Synapses. The Johns Hopkins University Press; Baltimore, MD: 2001.
-
- Hayashi T. Effects of sodium glutamate on the nervous system. Keio J. Med. 1954;3:183–192.
-
- Curtis DR, Phillis JW, Watkins JC. Chemical excitation of spinal neurons. Nature. 1959;183:611. - PubMed
-
- Sugiyama H, Ito I, Watanabe M. Glutamate receptor subtypes may be classified into two major categories: A study on Xenopus oocytes injected with rat brain mRNA. Neuron. 1989;3:129–132. - PubMed
-
- Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacological Rev. 1999;51:7–61. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

