Lipid droplets are novel sites of N-acylethanolamine inactivation by fatty acid amide hydrolase-2
- PMID: 19926788
- PMCID: PMC2807334
- DOI: 10.1074/jbc.M109.058461
Lipid droplets are novel sites of N-acylethanolamine inactivation by fatty acid amide hydrolase-2
Abstract
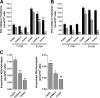
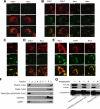
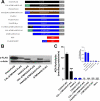
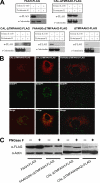
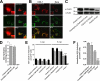
Anandamide (AEA) and other bioactive N-acylethanolamines (NAEs) are primarily inactivated by the enzyme fatty acid amide hydrolase (FAAH). Recently, FAAH-2 was discovered in humans, suggesting an additional enzyme can mediate NAE inactivation in higher mammals. Here, we performed a biochemical characterization of FAAH-2 and explored its capacity to hydrolyze NAEs in cells. In homogenate activity assays, FAAH-2 hydrolyzed AEA and palmitoylethanolamide (PEA) with activities approximately 6 and approximately 20% those of FAAH, respectively. In contrast, FAAH-2 hydrolyzed AEA and PEA in intact cells with rates approximately 30-40% those of FAAH, highlighting a potentially greater contribution toward NAE catabolism in vivo than previously appreciated. In contrast to endoplasmic reticulum-localized FAAH, immunofluorescence revealed FAAH-2 was localized on lipid droplets. Supporting this distribution pattern, the putative N-terminal hydrophobic region of FAAH-2 was identified as a functional lipid droplet localization sequence. Lipid droplet localization was essential for FAAH-2 activity as chimeras excluded from lipid droplets lacked activity and/or were poorly expressed. Lipid droplets represent novel sites of NAE inactivation. Therefore, we examined substrate delivery to these organelles. AEA was readily trafficked to lipid droplets, confirming that lipid droplets constitute functional sites of NAE inactivation. Collectively, these results establish FAAH-2 as a bone fide NAE-catabolizing enzyme and suggest that NAE inactivation is spatially separated in cells of higher mammals.
Figures
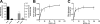
Similar articles
-
Identification of intracellular carriers for the endocannabinoid anandamide.Proc Natl Acad Sci U S A. 2009 Apr 14;106(15):6375-80. doi: 10.1073/pnas.0901515106. Epub 2009 Mar 23. Proc Natl Acad Sci U S A. 2009. PMID: 19307565 Free PMC article.
-
Endocannabinoid metabolism in the absence of fatty acid amide hydrolase (FAAH): discovery of phosphorylcholine derivatives of N-acyl ethanolamines.Biochemistry. 2006 Sep 26;45(38):11267-77. doi: 10.1021/bi061122s. Biochemistry. 2006. PMID: 16981687
-
N-acylethanolamine metabolism with special reference to N-acylethanolamine-hydrolyzing acid amidase (NAAA).Prog Lipid Res. 2010 Oct;49(4):299-315. doi: 10.1016/j.plipres.2010.02.003. Epub 2010 Feb 10. Prog Lipid Res. 2010. PMID: 20152858 Review.
-
N-acylethanolamine-hydrolyzing acid amidase and fatty acid amide hydrolase inhibition differentially affect N-acylethanolamine levels and macrophage activation.Biochim Biophys Acta Mol Cell Biol Lipids. 2017 May;1862(5):474-484. doi: 10.1016/j.bbalip.2017.01.001. Epub 2017 Jan 6. Biochim Biophys Acta Mol Cell Biol Lipids. 2017. PMID: 28065729
-
Fatty acid amide hydrolase inhibitors--progress and potential.CNS Neurol Disord Drug Targets. 2011 Aug;10(5):545-58. doi: 10.2174/187152711796234989. CNS Neurol Disord Drug Targets. 2011. PMID: 21631410 Review.
Cited by
-
Endocannabinoid signalling in innate and adaptive immunity.Immunology. 2015 Mar;144(3):352-364. doi: 10.1111/imm.12441. Immunology. 2015. PMID: 25585882 Free PMC article. Review.
-
Endocannabinoid signalling: has it got rhythm?Br J Pharmacol. 2010 Jun;160(3):530-43. doi: 10.1111/j.1476-5381.2010.00790.x. Br J Pharmacol. 2010. PMID: 20590563 Free PMC article. Review.
-
Abnormal accumulation of lipid droplets in neurons induces the conversion of alpha-Synuclein to proteolytic resistant forms in a Drosophila model of Parkinson's disease.PLoS Genet. 2021 Nov 17;17(11):e1009921. doi: 10.1371/journal.pgen.1009921. eCollection 2021 Nov. PLoS Genet. 2021. PMID: 34788284 Free PMC article.
-
The Endocannabinoid System as Pharmacological Target Derived from Its CNS Role in Energy Homeostasis and Reward. Applications in Eating Disorders and Addiction.Pharmaceuticals (Basel). 2011 Aug 10;4(8):1101-1136. doi: 10.3390/ph4081101. Pharmaceuticals (Basel). 2011. PMID: 32143540 Free PMC article. Review.
-
Lipid droplets and cellular lipid metabolism.Annu Rev Biochem. 2012;81:687-714. doi: 10.1146/annurev-biochem-061009-102430. Epub 2012 Apr 13. Annu Rev Biochem. 2012. PMID: 22524315 Free PMC article. Review.
References
-
- Lambert D. M., Fowler C. J. (2005) J. Med. Chem. 48, 5059–5087 - PubMed
-
- Walker J. M., Krey J. F., Chu C. J., Huang S. M. (2002) Chem. Phys. Lipids 121, 159–172 - PubMed
-
- Deutsch D. G., Chin S. A. (1993) Biochem. Pharmacol. 46, 791–796 - PubMed
-
- Cravatt B. F., Giang D. K., Mayfield S. P., Boger D. L., Lerner R. A., Gilula N. B. (1996) Nature 384, 83–87 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

