MOR23 promotes muscle regeneration and regulates cell adhesion and migration
- PMID: 19922870
- PMCID: PMC2780437
- DOI: 10.1016/j.devcel.2009.09.004
MOR23 promotes muscle regeneration and regulates cell adhesion and migration
Abstract
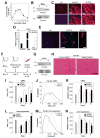
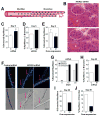
Odorant receptors (ORs) in the olfactory epithelium bind to volatile small molecules leading to the perception of smell. ORs are expressed in many tissues but their functions are largely unknown. We show multiple ORs display distinct mRNA expression patterns during myogenesis in vitro and muscle regeneration in vivo. Mouse OR23 (MOR23) expression is induced during muscle regeneration when muscle cells are extensively fusing and plays a key role in regulating migration and adhesion of muscle cells in vitro, two processes common during tissue repair. A soluble ligand for MOR23 is secreted by muscle cells in vitro and muscle tissue in vivo. MOR23 is necessary for proper skeletal muscle regeneration as loss of MOR23 leads to increased myofiber branching, commonly associated with muscular dystrophy. Together these data identify a functional role for an OR outside of the nose and suggest a larger role for ORs during tissue repair.
Figures

Similar articles
-
A new function for odorant receptors: MOR23 is necessary for normal tissue repair in skeletal muscle.Cell Adh Migr. 2010 Oct-Dec;4(4):502-6. doi: 10.4161/cam.4.4.12291. Cell Adh Migr. 2010. PMID: 20519965 Free PMC article.
-
Decrease of myofiber branching via muscle-specific expression of the olfactory receptor mOR23 in dystrophic muscle leads to protection against mechanical stress.Skelet Muscle. 2016 Jan 21;6:2. doi: 10.1186/s13395-016-0077-7. eCollection 2016. Skelet Muscle. 2016. PMID: 26798450 Free PMC article.
-
Olfactory receptors in non-chemosensory tissues.BMB Rep. 2012 Nov;45(11):612-22. doi: 10.5483/bmbrep.2012.45.11.232. BMB Rep. 2012. PMID: 23186999 Free PMC article. Review.
-
Regulation of Adipogenesis and Thermogenesis through Mouse Olfactory Receptor 23 Stimulated by α-Cedrene in 3T3-L1 Cells.Nutrients. 2018 Nov 16;10(11):1781. doi: 10.3390/nu10111781. Nutrients. 2018. PMID: 30453511 Free PMC article.
-
Noncoding RNAs in the regulation of skeletal muscle biology in health and disease.J Mol Med (Berl). 2016 Aug;94(8):853-66. doi: 10.1007/s00109-016-1443-y. Epub 2016 Jul 4. J Mol Med (Berl). 2016. PMID: 27377406 Free PMC article. Review.
Cited by
-
Olfactory Receptors and Tumorigenesis: Implications for Diagnosis and Targeted Therapy.Cell Biochem Biophys. 2024 Oct 4. doi: 10.1007/s12013-024-01556-7. Online ahead of print. Cell Biochem Biophys. 2024. PMID: 39365517 Review.
-
Analysis of single-cell transcriptomes links enrichment of olfactory receptors with cancer cell differentiation status and prognosis.Commun Biol. 2020 Sep 11;3(1):506. doi: 10.1038/s42003-020-01232-5. Commun Biol. 2020. PMID: 32917933 Free PMC article.
-
New insights into the epigenetic control of satellite cells.World J Stem Cells. 2015 Jul 26;7(6):945-55. doi: 10.4252/wjsc.v7.i6.945. World J Stem Cells. 2015. PMID: 26240681 Free PMC article. Review.
-
Key amino acids alter activity and trafficking of a well-conserved olfactory receptor.Am J Physiol Cell Physiol. 2022 Jun 1;322(6):C1279-C1288. doi: 10.1152/ajpcell.00440.2021. Epub 2022 May 11. Am J Physiol Cell Physiol. 2022. PMID: 35544696 Free PMC article.
-
G-protein coupled receptor BAI3 promotes myoblast fusion in vertebrates.Proc Natl Acad Sci U S A. 2014 Mar 11;111(10):3745-50. doi: 10.1073/pnas.1313886111. Epub 2014 Feb 24. Proc Natl Acad Sci U S A. 2014. PMID: 24567399 Free PMC article.
References
-
- Abbott KL, Loss JR, 2nd, Robida AM, Murphy TJ. Evidence that Galpha(q)-coupled receptor-induced interleukin-6 mRNA in vascular smooth muscle cells involves the nuclear factor of activated T cells. Mol Pharmacol. 2000;58:946–953. - PubMed
-
- Allen DL, Teitelbaum DH, Kurachi K. Growth factor stimulation of matrix metalloproteinase expression and myoblast migration and invasion in vitro. Am J Physiol Cell Physiol. 2003;284:C805–815. - PubMed
-
- Beggs ML, Nagarajan R, Taylor-Jones JM, Nolen G, Macnicol M, Peterson CA. Alterations in the TGFbeta signaling pathway in myogenic progenitors with age. Aging Cell. 2004;3:353–361. - PubMed
-
- Bondesen BA, Mills ST, Kegley KM, Pavlath GK. The COX-2 pathway is essential during early stages of skeletal muscle regeneration. Am J Physiol Cell Physiol. 2004;287:C475–483. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR052730/AR/NIAMS NIH HHS/United States
- T32-GM08367/GM/NIGMS NIH HHS/United States
- R01 AR052730-03/AR/NIAMS NIH HHS/United States
- AR-052730/AR/NIAMS NIH HHS/United States
- R01 AR047314/AR/NIAMS NIH HHS/United States
- AR-051372/AR/NIAMS NIH HHS/United States
- T32 GM008367/GM/NIGMS NIH HHS/United States
- R01 AR052730-02/AR/NIAMS NIH HHS/United States
- R01 AR051372/AR/NIAMS NIH HHS/United States
- AR-047314/AR/NIAMS NIH HHS/United States
- R01 AR052730-04/AR/NIAMS NIH HHS/United States
- R01 AR052730-01A1/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

