Targeted KRAS mutation assessment on patient tumor histologic material in real time diagnostics
- PMID: 19888477
- PMCID: PMC2768905
- DOI: 10.1371/journal.pone.0007746
Targeted KRAS mutation assessment on patient tumor histologic material in real time diagnostics
Abstract
Background: Testing for tumor specific mutations on routine formalin-fixed paraffin-embedded (FFPE) tissues may predict response to treatment in Medical Oncology and has already entered diagnostics, with KRAS mutation assessment as a paradigm. The highly sensitive real time PCR (Q-PCR) methods developed for this purpose are usually standardized under optimal template conditions. In routine diagnostics, however, suboptimal templates pose the challenge. Herein, we addressed the applicability of sequencing and two Q-PCR methods on prospectively assessed diagnostic cases for KRAS mutations.
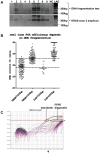
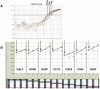
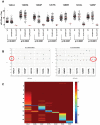
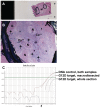
Methodology/principal findings: Tumor FFPE-DNA from 135 diagnostic and 75 low-quality control samples was obtained upon macrodissection, tested for fragmentation and assessed for KRAS mutations with dideoxy-sequencing and with two Q-PCR methods (Taqman-minor-groove-binder [TMGB] probes and DxS-KRAS-IVD). Samples with relatively well preserved DNA could be accurately analyzed with sequencing, while Q-PCR methods yielded informative results even in cases with very fragmented DNA (p<0.0001) with 100% sensitivity and specificity vs each other. However, Q-PCR efficiency (Ct values) also depended on DNA-fragmentation (p<0.0001). Q-PCR methods were sensitive to detect<or=1% mutant cells, provided that samples yielded cycle thresholds (Ct)<29, but this condition was met in only 38.5% of diagnostic samples. In comparison, FFPE samples (>99%) could accurately be analyzed at a sensitivity level of 10% (external validation of TMGB results). DNA quality and tumor cell content were the main reasons for discrepant sequencing/Q-PCR results (1.5%).
Conclusions/significance: Diagnostic targeted mutation assessment on FFPE-DNA is very efficient with Q-PCR methods in comparison to dideoxy-sequencing. However, DNA fragmentation/amplification capacity and tumor DNA content must be considered for the interpretation of Q-PCR results in order to provide accurate information for clinical decision making.
Conflict of interest statement
Figures

Similar articles
-
Validation of targeted next-generation sequencing for RAS mutation detection in FFPE colorectal cancer tissues: comparison with Sanger sequencing and ARMS-Scorpion real-time PCR.BMJ Open. 2016 Jan 8;6(1):e009532. doi: 10.1136/bmjopen-2015-009532. BMJ Open. 2016. PMID: 26747035 Free PMC article.
-
Detection of KRAS mutations in colorectal carcinoma patients with an integrated PCR/sequencing and real-time PCR approach.Pharmacogenomics. 2010 Aug;11(8):1169-79. doi: 10.2217/pgs.10.86. Pharmacogenomics. 2010. PMID: 20712532 Clinical Trial.
-
KRAS and BRAF mutation analysis in routine molecular diagnostics: comparison of three testing methods on formalin-fixed, paraffin-embedded tumor-derived DNA.J Mol Diagn. 2012 May-Jun;14(3):247-55. doi: 10.1016/j.jmoldx.2012.01.011. Epub 2012 Mar 14. J Mol Diagn. 2012. PMID: 22425762
-
Competitive allele specific TaqMan PCR for KRAS, BRAF and EGFR mutation detection in clinical formalin fixed paraffin embedded samples.Exp Mol Pathol. 2012 Jun;92(3):275-80. doi: 10.1016/j.yexmp.2012.03.001. Epub 2012 Mar 7. Exp Mol Pathol. 2012. PMID: 22426079
-
[Comparative analysis of real-time quantitative PCR-Sanger sequencing method and TaqMan probe method for detection of KRAS/BRAF mutation in colorectal carcinomas].Zhonghua Bing Li Xue Za Zhi. 2014 Feb;43(2):77-82. Zhonghua Bing Li Xue Za Zhi. 2014. PMID: 24742565 Chinese.
Cited by
-
Accurate Detection of Hepatitis B Virus G1896A Mutant by Developed Taqman-ARMS Followed a Strict Control System.J Clin Lab Anal. 2016 Jul;30(4):315-8. doi: 10.1002/jcla.21857. Epub 2015 May 13. J Clin Lab Anal. 2016. PMID: 25968238 Free PMC article.
-
TP53 mutations and protein immunopositivity may predict for poor outcome but also for trastuzumab benefit in patients with early breast cancer treated in the adjuvant setting.Oncotarget. 2016 May 31;7(22):32731-53. doi: 10.18632/oncotarget.9022. Oncotarget. 2016. PMID: 27129168 Free PMC article.
-
KRAS and BRAF mutation analysis in colorectal adenocarcinoma specimens with a low percentage of tumor cells.Mol Diagn Ther. 2013 Jun;17(3):193-203. doi: 10.1007/s40291-013-0025-8. Mol Diagn Ther. 2013. PMID: 23606169 Free PMC article.
-
Performance and cost efficiency of KRAS mutation testing for metastatic colorectal cancer in routine diagnosis: the MOKAECM study, a nationwide experience.PLoS One. 2013 Jul 25;8(7):e68945. doi: 10.1371/journal.pone.0068945. Print 2013. PLoS One. 2013. PMID: 23935912 Free PMC article.
-
Detection of hepatitis B virus A1762T/G1764A mutant by amplification refractory mutation system.Braz J Infect Dis. 2014 May-Jun;18(3):261-5. doi: 10.1016/j.bjid.2013.09.005. Epub 2014 Jan 3. Braz J Infect Dis. 2014. PMID: 24389280 Free PMC article.
References
-
- Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–1174. - PubMed
-
- Stewart DJ, Kurzrock R. Cancer: the road to Amiens. J Clin Oncol. 2009;27:328–333. - PubMed
-
- Messersmith WA, Ahnen DJ. Targeting EGFR in colorectal cancer. N Engl J Med. 2008;359:1834–1836. - PubMed
-
- Bach PB. Limits on Medicare's ability to control rising spending on cancer drugs. N Engl J Med. 2009;360:626–633. - PubMed
-
- EMEA. EMEA/405113/2007. 2007.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

