Modulation of LMP2A expression by a newly identified Epstein-Barr virus-encoded microRNA miR-BART22
- PMID: 19881953
- PMCID: PMC2767219
- DOI: 10.1593/neo.09888
Modulation of LMP2A expression by a newly identified Epstein-Barr virus-encoded microRNA miR-BART22
Abstract
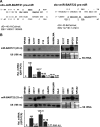
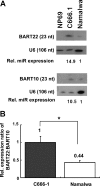
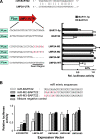
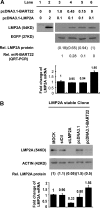
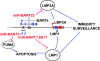
Infection with the Epstein-Barr virus (EBV) is a strong predisposing factor in the development of nasopharyngeal carcinoma (NPC). Many viral gene products including EBNA1, LMP1, and LMP2 have been implicated in NPC tumorigenesis, although the de novo control of these viral oncoproteins remains largely unclear. The recent discovery of EBV-encoded viral microRNA (miRNA) in lymphoid malignancies has prompted us to examine the NPC-associated EBV miRNA. Using large-scale cloning analysis on EBV-positive NPC cells, two novel EBV miRNA, now named miR-BART21 and miR-BART22, were identified. These two EBV-encoded miRNA are abundantly expressed in most NPC samples. We found two nucleotide variations in the primary transcript of miR-BART22, which we experimentally confirmed to augment its biogenesis in vitro and thus may underline the high and consistent expression of miR-BART22 in NPC tumors. More importantly, we determined that the EBV latent membrane protein 2A (LMP2A) is the putative target of miR-BART22. LMP2A is a potent immunogenic viral antigen that is recognized by the cytotoxic T cells; down-modulation of LMP2A expression by miR-BART22 may permit escape of EBV-infected cells from host immune surveillance. Taken together, we demonstrated that two newly identified EBV-encoded miRNA are highly expressed in NPC. Specific sequence variations on the prevalent EBV strain in our locality might contribute to the higher miR-BART22 expression level in our NPC samples. Our findings emphasize the role of miR-BART22 in modulating LMP2A expression, which may facilitate NPC carcinogenesis by evading the host immune response.
Figures

Similar articles
-
[Relationship between MAP3K5 and Epstein-Barr virus-encoded miR-BART22 expression in nasopharyngeal carcinoma].Nan Fang Yi Ke Da Xue Xue Bao. 2011 Jun;31(7):1146-9. Nan Fang Yi Ke Da Xue Xue Bao. 2011. PMID: 21764682 Chinese.
-
Epstein-Barr virus-encoded LMP2A regulates viral and cellular gene expression by modulation of the NF-kappaB transcription factor pathway.Proc Natl Acad Sci U S A. 2004 Nov 2;101(44):15730-5. doi: 10.1073/pnas.0402135101. Epub 2004 Oct 21. Proc Natl Acad Sci U S A. 2004. PMID: 15498875 Free PMC article.
-
Cinobufotalin powerfully reversed EBV-miR-BART22-induced cisplatin resistance via stimulating MAP2K4 to antagonize non-muscle myosin heavy chain IIA/glycogen synthase 3β/β-catenin signaling pathway.EBioMedicine. 2019 Oct;48:386-404. doi: 10.1016/j.ebiom.2019.08.040. Epub 2019 Oct 5. EBioMedicine. 2019. PMID: 31594754 Free PMC article.
-
Pathogenic role of Epstein-Barr virus latent membrane protein-1 in the development of nasopharyngeal carcinoma.Cancer Lett. 2013 Aug 28;337(1):1-7. doi: 10.1016/j.canlet.2013.05.018. Epub 2013 May 17. Cancer Lett. 2013. PMID: 23689138 Review.
-
Emerging roles of small Epstein-Barr virus derived non-coding RNAs in epithelial malignancy.Int J Mol Sci. 2013 Aug 23;14(9):17378-409. doi: 10.3390/ijms140917378. Int J Mol Sci. 2013. PMID: 23979421 Free PMC article. Review.
Cited by
-
EBV-driven B-cell lymphoproliferative disorders: from biology, classification and differential diagnosis to clinical management.Exp Mol Med. 2015 Jan 23;47(1):e132. doi: 10.1038/emm.2014.82. Exp Mol Med. 2015. PMID: 25613729 Free PMC article. Review.
-
Cross-Kingdom Interaction of miRNAs and Gut Microbiota with Non-Invasive Diagnostic and Therapeutic Implications in Colorectal Cancer.Int J Mol Sci. 2023 Jun 23;24(13):10520. doi: 10.3390/ijms241310520. Int J Mol Sci. 2023. PMID: 37445698 Free PMC article. Review.
-
Characterization of rare transforming KRAS mutations in sporadic colorectal cancer.Cancer Biol Ther. 2014 Jun 1;15(6):768-76. doi: 10.4161/cbt.28550. Epub 2014 Mar 18. Cancer Biol Ther. 2014. PMID: 24642870 Free PMC article.
-
Integrated mRNA and microRNA transcriptome sequencing characterizes sequence variants and mRNA-microRNA regulatory network in nasopharyngeal carcinoma model systems.FEBS Open Bio. 2014 Jan 13;4:128-40. doi: 10.1016/j.fob.2014.01.004. eCollection 2014. FEBS Open Bio. 2014. PMID: 24490137 Free PMC article.
-
MicroRNAs of Epstein-Barr Virus Control Innate and Adaptive Antiviral Immunity.J Virol. 2017 Jul 27;91(16):e01667-16. doi: 10.1128/JVI.01667-16. Print 2017 Aug 15. J Virol. 2017. PMID: 28592533 Free PMC article. Review.
References
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Lo KW, To KF, Huang DP. Focus on nasopharyngeal carcinoma. Cancer Cell. 2004;5:423–428. - PubMed
-
- Raab-Traub N. Epstein-Barr virus in the pathogenesis of NPC. Semin Cancer Biol. 2002;12:431–441. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
