The role of DNA shape in protein-DNA recognition
- PMID: 19865164
- PMCID: PMC2793086
- DOI: 10.1038/nature08473
The role of DNA shape in protein-DNA recognition
Abstract
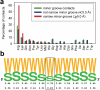
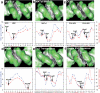
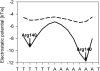
The recognition of specific DNA sequences by proteins is thought to depend on two types of mechanism: one that involves the formation of hydrogen bonds with specific bases, primarily in the major groove, and one involving sequence-dependent deformations of the DNA helix. By comprehensively analysing the three-dimensional structures of protein-DNA complexes, here we show that the binding of arginine residues to narrow minor grooves is a widely used mode for protein-DNA recognition. This readout mechanism exploits the phenomenon that narrow minor grooves strongly enhance the negative electrostatic potential of the DNA. The nucleosome core particle offers a prominent example of this effect. Minor-groove narrowing is often associated with the presence of A-tracts, AT-rich sequences that exclude the flexible TpA step. These findings indicate that the ability to detect local variations in DNA shape and electrostatic potential is a general mechanism that enables proteins to use information in the minor groove, which otherwise offers few opportunities for the formation of base-specific hydrogen bonds, to achieve DNA-binding specificity.
Figures


Comment in
-
Structural biology: DNA binding shapes up.Nature. 2009 Oct 29;461(7268):1225-6. doi: 10.1038/4611225a. Nature. 2009. PMID: 19865161 No abstract available.
Similar articles
-
Electrostatic interactions between arginines and the minor groove in the nucleosome.J Biomol Struct Dyn. 2010 Jun;27(6):861-6. doi: 10.1080/07391102.2010.10508587. J Biomol Struct Dyn. 2010. PMID: 20232938 Free PMC article.
-
Sequence-dependent Kink-and-Slide deformations of nucleosomal DNA facilitated by histone arginines bound in the minor groove.J Biomol Struct Dyn. 2010 Jun;27(6):843-59. doi: 10.1080/07391102.2010.10508586. J Biomol Struct Dyn. 2010. PMID: 20232937 Free PMC article.
-
Origins of specificity in protein-DNA recognition.Annu Rev Biochem. 2010;79:233-69. doi: 10.1146/annurev-biochem-060408-091030. Annu Rev Biochem. 2010. PMID: 20334529 Free PMC article. Review.
-
Recognition of 5'-YpG-3' sequences by coupled stacking/hydrogen bonding interactions with amino acid residues.J Mol Biol. 2004 Jan 9;335(2):399-408. doi: 10.1016/j.jmb.2003.10.071. J Mol Biol. 2004. PMID: 14672650
-
What drives proteins into the major or minor grooves of DNA?J Mol Biol. 2007 Jan 5;365(1):1-9. doi: 10.1016/j.jmb.2006.09.059. Epub 2006 Sep 27. J Mol Biol. 2007. PMID: 17055530 Free PMC article. Review.
Cited by
-
Utilizing biological experimental data and molecular dynamics for the classification of mutational hotspots through machine learning.Bioinform Adv. 2024 Aug 26;4(1):vbae125. doi: 10.1093/bioadv/vbae125. eCollection 2024. Bioinform Adv. 2024. PMID: 39239360 Free PMC article.
-
Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution.Proc Natl Acad Sci U S A. 2016 Aug 9;113(32):9057-62. doi: 10.1073/pnas.1606667113. Epub 2016 Jul 25. Proc Natl Acad Sci U S A. 2016. PMID: 27457959 Free PMC article.
-
Crystal structure of the DNA-bound VapBC2 antitoxin/toxin pair from Rickettsia felis.Nucleic Acids Res. 2012 Apr;40(7):3245-58. doi: 10.1093/nar/gkr1167. Epub 2011 Dec 2. Nucleic Acids Res. 2012. PMID: 22140099 Free PMC article.
-
Quantitative modeling of transcription factor binding specificities using DNA shape.Proc Natl Acad Sci U S A. 2015 Apr 14;112(15):4654-9. doi: 10.1073/pnas.1422023112. Epub 2015 Mar 9. Proc Natl Acad Sci U S A. 2015. PMID: 25775564 Free PMC article.
-
Systematic comparison and prediction of the effects of missense mutations on protein-DNA and protein-RNA interactions.PLoS Comput Biol. 2021 Apr 19;17(4):e1008951. doi: 10.1371/journal.pcbi.1008951. eCollection 2021 Apr. PLoS Comput Biol. 2021. PMID: 33872313 Free PMC article.
References
-
- Garvie CW, Wolberger C. Recognition of specific DNA sequences. Mol Cell. 2001;8(5):937–946. - PubMed
-
- Travers AA. DNA conformation and protein binding. Annu Rev Biochem. 1989;58:427–452. - PubMed
-
- Shakked Z, et al. Determinants of repressor/operator recognition from the structure of the trp operator binding site. Nature. 1994;368(6470):469–473. - PubMed
-
- Lu XJ, Shakked Z, Olson WK. A-form conformational motifs in ligand-bound DNA structures. J Mol Biol. 2000;300(4):819–840. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

