Crystal structure of a "nonfoldable" insulin: impaired folding efficiency despite native activity
- PMID: 19850922
- PMCID: PMC2787385
- DOI: 10.1074/jbc.M109.046888
Crystal structure of a "nonfoldable" insulin: impaired folding efficiency despite native activity
Abstract
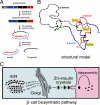
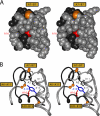
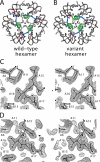
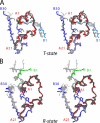
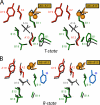
Protein evolution is constrained by folding efficiency ("foldability") and the implicit threat of toxic misfolding. A model is provided by proinsulin, whose misfolding is associated with beta-cell dysfunction and diabetes mellitus. An insulin analogue containing a subtle core substitution (Leu(A16) --> Val) is biologically active, and its crystal structure recapitulates that of the wild-type protein. As a seeming paradox, however, Val(A16) blocks both insulin chain combination and the in vitro refolding of proinsulin. Disulfide pairing in mammalian cell culture is likewise inefficient, leading to misfolding, endoplasmic reticular stress, and proteosome-mediated degradation. Val(A16) destabilizes the native state and so presumably perturbs a partial fold that directs initial disulfide pairing. Substitutions elsewhere in the core similarly destabilize the native state but, unlike Val(A16), preserve folding efficiency. We propose that Leu(A16) stabilizes nonlocal interactions between nascent alpha-helices in the A- and B-domains to facilitate initial pairing of Cys(A20) and Cys(B19), thus surmounting their wide separation in sequence. Although Val(A16) is likely to destabilize this proto-core, its structural effects are mitigated once folding is achieved. Classical studies of insulin chain combination in vitro have illuminated the impact of off-pathway reactions on the efficiency of native disulfide pairing. The capability of a polypeptide sequence to fold within the endoplasmic reticulum may likewise be influenced by kinetic or thermodynamic partitioning among on- and off-pathway disulfide intermediates. The properties of [Val(A16)]insulin and [Val(A16)]proinsulin demonstrate that essential contributions of conserved residues to folding may be inapparent once the native state is achieved.
Figures

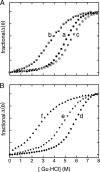
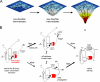
 ). B, corresponding studies of DKP-insulin analogues: parent monomer (d, ●), GlyA2 analogue (e, ▾), and SerA6,SerA11 two-disulfide analogue (f, ▴). Fit to a two-state model by nonlinear least squares regression was in each case characterized by R-factors greater than 0.99, justifying extrapolation of free energies (ΔGu) to zero denaturant concentration (
). B, corresponding studies of DKP-insulin analogues: parent monomer (d, ●), GlyA2 analogue (e, ▾), and SerA6,SerA11 two-disulfide analogue (f, ▴). Fit to a two-state model by nonlinear least squares regression was in each case characterized by R-factors greater than 0.99, justifying extrapolation of free energies (ΔGu) to zero denaturant concentration (
Similar articles
-
Structural Lessons From the Mutant Proinsulin Syndrome.Front Endocrinol (Lausanne). 2021 Sep 30;12:754693. doi: 10.3389/fendo.2021.754693. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 34659132 Free PMC article. Review.
-
Peptide Model of the Mutant Proinsulin Syndrome. II. Nascent Structure and Biological Implications.Front Endocrinol (Lausanne). 2022 Mar 1;13:821091. doi: 10.3389/fendo.2022.821091. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35299958 Free PMC article.
-
Evolution of insulin at the edge of foldability and its medical implications.Proc Natl Acad Sci U S A. 2020 Nov 24;117(47):29618-29628. doi: 10.1073/pnas.2010908117. Epub 2020 Nov 5. Proc Natl Acad Sci U S A. 2020. PMID: 33154160 Free PMC article.
-
Chiral mutagenesis of insulin. Foldability and function are inversely regulated by a stereospecific switch in the B chain.Biochemistry. 2005 Apr 5;44(13):4984-99. doi: 10.1021/bi048025o. Biochemistry. 2005. PMID: 15794637 Free PMC article.
-
Diabetes-Associated Mutations in Proinsulin Provide a "Molecular Rheostat" of Nascent Foldability.Curr Diab Rep. 2022 Feb;22(2):85-94. doi: 10.1007/s11892-022-01447-2. Epub 2022 Feb 4. Curr Diab Rep. 2022. PMID: 35119630 Review.
Cited by
-
Structural Lessons From the Mutant Proinsulin Syndrome.Front Endocrinol (Lausanne). 2021 Sep 30;12:754693. doi: 10.3389/fendo.2021.754693. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 34659132 Free PMC article. Review.
-
Structure-based stabilization of insulin as a therapeutic protein assembly via enhanced aromatic-aromatic interactions.J Biol Chem. 2018 Jul 13;293(28):10895-10910. doi: 10.1074/jbc.RA118.003650. Epub 2018 Jun 7. J Biol Chem. 2018. PMID: 29880646 Free PMC article.
-
Deciphering a molecular mechanism of neonatal diabetes mellitus by the chemical synthesis of a protein diastereomer, [D-AlaB8]human proinsulin.J Biol Chem. 2014 Aug 22;289(34):23683-92. doi: 10.1074/jbc.M114.572040. Epub 2014 Jul 7. J Biol Chem. 2014. PMID: 25002580 Free PMC article.
-
A Polymorphism in the Gene Encoding Heat Shock Factor 1 (HSF1) Increases the Risk of Type 2 Diabetes: A Pilot Study Supports a Role for Impaired Protein Folding in Disease Pathogenesis.Life (Basel). 2022 Nov 20;12(11):1936. doi: 10.3390/life12111936. Life (Basel). 2022. PMID: 36431071 Free PMC article.
-
INS-gene mutations: from genetics and beta cell biology to clinical disease.Mol Aspects Med. 2015 Apr;42:3-18. doi: 10.1016/j.mam.2014.12.001. Epub 2014 Dec 24. Mol Aspects Med. 2015. PMID: 25542748 Free PMC article. Review.
References
-
- Onuchic J. N., Luthey-Schulten Z., Wolynes P. G. (1997) Annu. Rev. Phys. Chem. 48, 545–600 - PubMed
-
- Dobson C. M., Karplus M. (1999) Curr. Opin. Struct. Biol. 9, 92–101 - PubMed
-
- Shakhnovich E., Farztdinov G., Gutin A. M., Karplus M. (1991) Phys. Rev. Lett. 67, 1665–1668 - PubMed
-
- Onuchic J. N., Socci N. D., Luthey-Schulten Z., Wolynes P. G. (1996) Fold. Des. 1, 441–450 - PubMed
-
- Dill K. A., Chan H. S. (1997) Nat. Struct. Biol. 4, 10–19 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

