Murine hepatitis virus nonstructural protein 4 regulates virus-induced membrane modifications and replication complex function
- PMID: 19846526
- PMCID: PMC2798404
- DOI: 10.1128/JVI.01772-09
Murine hepatitis virus nonstructural protein 4 regulates virus-induced membrane modifications and replication complex function
Abstract
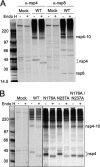
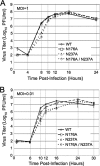
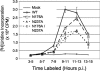
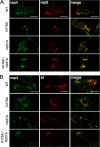
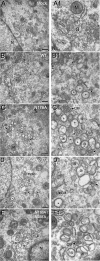
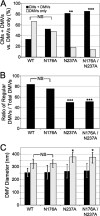
Positive-strand RNA viruses induce modifications of cytoplasmic membranes to form replication complexes. For coronaviruses, replicase nonstructural protein 4 (nsp4) has been proposed to function in the formation and organization of replication complexes. Murine hepatitis virus (MHV) nsp4 is glycosylated at residues Asn176 (N176) and N237 during plasmid expression of nsp4 in cells. To test if MHV nsp4 residues N176 and N237 are glycosylated during virus replication and to determine the effects of N176 and N237 on nsp4 function and MHV replication, alanine substitutions of nsp4 N176, N237, or both were engineered into the MHV-A59 genome. The N176A, N237A, and N176A/N237A mutant viruses were viable, and N176 and N237 were glycosylated during infection of wild-type (wt) and mutant viruses. The nsp4 glycosylation mutants exhibited impaired virus growth and RNA synthesis, with the N237A and N176A/N237A mutant viruses demonstrating more profound defects in virus growth and RNA synthesis. Electron microscopic analysis of ultrastructure from infected cells demonstrated that the nsp4 mutants had aberrant morphology of virus-induced double-membrane vesicles (DMVs) compared to those infected with wt virus. The degree of altered DMV morphology directly correlated with the extent of impairment in viral RNA synthesis and virus growth of the nsp4 mutant viruses. The results indicate that nsp4 plays a critical role in the organization and stability of DMVs. The results also support the conclusion that the structure of DMVs is essential for efficient RNA synthesis and optimal replication of coronaviruses.
Figures

Similar articles
-
Mutations across murine hepatitis virus nsp4 alter virus fitness and membrane modifications.J Virol. 2015 Feb;89(4):2080-9. doi: 10.1128/JVI.02776-14. Epub 2014 Dec 3. J Virol. 2015. PMID: 25473044 Free PMC article.
-
Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles.Virology. 2008 May 25;375(1):118-29. doi: 10.1016/j.virol.2008.01.018. Epub 2008 Mar 4. Virology. 2008. PMID: 18295294 Free PMC article.
-
Genetic analysis of Murine hepatitis virus nsp4 in virus replication.J Virol. 2007 Nov;81(22):12554-63. doi: 10.1128/JVI.01257-07. Epub 2007 Sep 12. J Virol. 2007. PMID: 17855548 Free PMC article.
-
The double-membrane vesicle (DMV): a virus-induced organelle dedicated to the replication of SARS-CoV-2 and other positive-sense single-stranded RNA viruses.Cell Mol Life Sci. 2022 Jul 16;79(8):425. doi: 10.1007/s00018-022-04469-x. Cell Mol Life Sci. 2022. PMID: 35841484 Free PMC article. Review.
-
Biogenesis and architecture of arterivirus replication organelles.Virus Res. 2016 Jul 15;220:70-90. doi: 10.1016/j.virusres.2016.04.001. Epub 2016 Apr 9. Virus Res. 2016. PMID: 27071852 Free PMC article. Review.
Cited by
-
Coronaviruses: an overview of their replication and pathogenesis.Methods Mol Biol. 2015;1282:1-23. doi: 10.1007/978-1-4939-2438-7_1. Methods Mol Biol. 2015. PMID: 25720466 Free PMC article. Review.
-
Murine hepatitis virus nsp4 N258T mutants are not temperature-sensitive.Virology. 2013 Jan 20;435(2):210-3. doi: 10.1016/j.virol.2012.10.001. Epub 2012 Oct 23. Virology. 2013. PMID: 23099203 Free PMC article.
-
New insights on the role of paired membrane structures in coronavirus replication.Virus Res. 2015 Apr 16;202:33-40. doi: 10.1016/j.virusres.2014.12.021. Epub 2014 Dec 27. Virus Res. 2015. PMID: 25550072 Free PMC article.
-
Post-translational modifications of coronavirus proteins: roles and function.Future Virol. 2018 Jun;13(6):405-430. doi: 10.2217/fvl-2018-0008. Epub 2018 May 21. Future Virol. 2018. PMID: 32201497 Free PMC article. Review.
-
Aminoterminal amphipathic α-helix AH1 of hepatitis C virus nonstructural protein 4B possesses a dual role in RNA replication and virus production.PLoS Pathog. 2014 Nov 13;10(10):e1004501. doi: 10.1371/journal.ppat.1004501. eCollection 2014 Oct. PLoS Pathog. 2014. PMID: 25392992 Free PMC article.
References
-
- Bredenbeek, P. J., C. J. Pachuk, A. F. H. Noten, J. Charite, W. Luytjes, S. R. Weiss, and W. J. M. Spaan. 1990. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 18:1825-1832. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

