Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer
- PMID: 19843336
- PMCID: PMC2770572
- DOI: 10.1186/1471-2407-9-374
Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer
Abstract
Background: MicroRNAs (MiRNAs) are short non-coding RNAs that control protein expression through various mechanisms. Their altered expression has been shown to be associated with various cancers. The aim of this study was to profile miRNA expression in colorectal cancer (CRC) and to analyze the function of specific miRNAs in CRC cells. MirVana miRNA Bioarrays were used to determine the miRNA expression profile in eight CRC cell line models, 45 human CRC samples of different stages, and four matched normal colon tissue samples. SW620 CRC cells were stably transduced with miR-143 or miR-145 expression vectors and analyzed in vitro for cell proliferation, cell differentiation and anchorage-independent growth. Signalling pathways associated with differentially expressed miRNAs were identified using a gene set enrichment analysis.
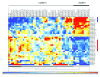
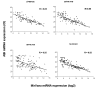
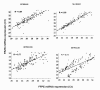
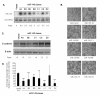
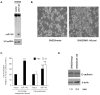
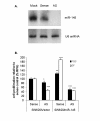
Results: The expression analysis of clinical CRC samples identified 37 miRNAs that were differentially expressed between CRC and normal tissue. Furthermore, several of these miRNAs were associated with CRC tumor progression including loss of miR-133a and gain of miR-224. We identified 11 common miRNAs that were differentially expressed between normal colon and CRC in both the cell line models and clinical samples. In vitro functional studies indicated that miR-143 and miR-145 appear to function in opposing manners to either inhibit or augment cell proliferation in a metastatic CRC model. The pathways targeted by miR-143 and miR-145 showed no significant overlap. Furthermore, gene expression analysis of metastatic versus non-metastatic isogenic cell lines indicated that miR-145 targets involved in cell cycle and neuregulin pathways were significantly down-regulated in the metastatic context.
Conclusion: MiRNAs showing altered expression at different stages of CRC could be targets for CRC therapies and be further developed as potential diagnostic and prognostic analytes. The identified biological processes and signalling pathways collectively targeted by co-expressed miRNAs in CRC provide a basis for understanding the functional role of miRNAs in cancer.
Figures

Similar articles
-
Functional screening identifies miRNAs influencing apoptosis and proliferation in colorectal cancer.PLoS One. 2014 Jun 3;9(6):e96767. doi: 10.1371/journal.pone.0096767. eCollection 2014. PLoS One. 2014. PMID: 24892549 Free PMC article.
-
Association of differential miRNA expression with hepatic vs. peritoneal metastatic spread in colorectal cancer.BMC Cancer. 2018 Feb 20;18(1):201. doi: 10.1186/s12885-018-4043-0. BMC Cancer. 2018. PMID: 29463215 Free PMC article.
-
The expression of microRNA-375 in plasma and tissue is matched in human colorectal cancer.BMC Cancer. 2014 Sep 25;14:714. doi: 10.1186/1471-2407-14-714. BMC Cancer. 2014. PMID: 25255814 Free PMC article.
-
The role of microRNA in metastatic colorectal cancer and its significance in cancer prognosis and treatment.Acta Biochim Pol. 2012;59(4):467-74. Epub 2012 Nov 21. Acta Biochim Pol. 2012. PMID: 23173124 Review.
-
miRNA Clusters with Down-Regulated Expression in Human Colorectal Cancer and Their Regulation.Int J Mol Sci. 2020 Jun 29;21(13):4633. doi: 10.3390/ijms21134633. Int J Mol Sci. 2020. PMID: 32610706 Free PMC article. Review.
Cited by
-
Circulating Serum miRNAs as Diagnostic Markers for Colorectal Cancer.PLoS One. 2016 May 2;11(5):e0154130. doi: 10.1371/journal.pone.0154130. eCollection 2016. PLoS One. 2016. PMID: 27135244 Free PMC article.
-
Environmental chemical exposures and human epigenetics.Int J Epidemiol. 2012 Feb;41(1):79-105. doi: 10.1093/ije/dyr154. Epub 2011 Dec 13. Int J Epidemiol. 2012. PMID: 22253299 Free PMC article.
-
Identification of a two-layer regulatory network of proliferation-related microRNAs in hepatoma cells.Nucleic Acids Res. 2012 Nov 1;40(20):10478-93. doi: 10.1093/nar/gks789. Epub 2012 Aug 25. Nucleic Acids Res. 2012. PMID: 22923518 Free PMC article.
-
Power in pairs: assessing the statistical value of paired samples in tests for differential expression.BMC Genomics. 2018 Dec 20;19(1):953. doi: 10.1186/s12864-018-5236-2. BMC Genomics. 2018. PMID: 30572829 Free PMC article.
-
Differential expression of microRNAs in tumors from chronically inflamed or genetic (APC(Min/+)) models of colon cancer.PLoS One. 2011 Apr 12;6(4):e18501. doi: 10.1371/journal.pone.0018501. PLoS One. 2011. PMID: 21532750 Free PMC article.
References
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

