Targeting proteins for degradation
- PMID: 19841631
- PMCID: PMC4228941
- DOI: 10.1038/nchembio.250
Targeting proteins for degradation
Erratum in
- Nat Chem Biol. 2009 Dec;5(12):954
Abstract
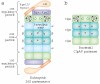
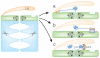
Protein degradation plays a central role in many cellular functions. Misfolded and damaged proteins are removed from the cell to avoid toxicity. The concentrations of regulatory proteins are adjusted by degradation at the appropriate time. Both foreign and native proteins are digested into small peptides as part of the adaptive immune response. In eukaryotic cells, an ATP-dependent protease called the proteasome is responsible for much of this proteolysis. Proteins are targeted for proteasomal degradation by a two-part degron, which consists of a proteasome binding signal and a degradation initiation site. Here we describe how both components contribute to the specificity of degradation.
Figures

Similar articles
-
ATP-Dependent inactivation and sequestration of ornithine decarboxylase by the 26S proteasome are prerequisites for degradation.Mol Cell Biol. 1999 Oct;19(10):7216-27. doi: 10.1128/MCB.19.10.7216. Mol Cell Biol. 1999. PMID: 10490656 Free PMC article.
-
Identification of Caspase-6-mediated processing of the valosin containing protein (p97) in Alzheimer's disease: a novel link to dysfunction in ubiquitin proteasome system-mediated protein degradation.J Neurosci. 2010 Apr 28;30(17):6132-42. doi: 10.1523/JNEUROSCI.5874-09.2010. J Neurosci. 2010. PMID: 20427671 Free PMC article.
-
Identification of the Cdc48•20S proteasome as an ancient AAA+ proteolytic machine.Science. 2012 Aug 17;337(6096):843-6. doi: 10.1126/science.1224352. Epub 2012 Jul 26. Science. 2012. PMID: 22837385 Free PMC article.
-
Ubiquitin recognition by the proteasome.J Biochem. 2017 Feb 1;161(2):113-124. doi: 10.1093/jb/mvw091. J Biochem. 2017. PMID: 28069863 Review.
-
Recognition of Client Proteins by the Proteasome.Annu Rev Biophys. 2017 May 22;46:149-173. doi: 10.1146/annurev-biophys-070816-033719. Epub 2017 Mar 9. Annu Rev Biophys. 2017. PMID: 28301771 Review.
Cited by
-
Epidermal growth factor promotes protein degradation of epithelial protein lost in neoplasm (EPLIN), a putative metastasis suppressor, during epithelial-mesenchymal transition.J Biol Chem. 2013 Jan 18;288(3):1469-79. doi: 10.1074/jbc.M112.438341. Epub 2012 Nov 27. J Biol Chem. 2013. PMID: 23188829 Free PMC article.
-
Myc proteins in brain tumor development and maintenance.Ups J Med Sci. 2012 May;117(2):122-31. doi: 10.3109/03009734.2012.658975. Epub 2012 Feb 21. Ups J Med Sci. 2012. PMID: 22348395 Free PMC article.
-
Inducible Rpt3, a Proteasome Component, Knockout in Adult Skeletal Muscle Results in Muscle Atrophy.Front Cell Dev Biol. 2020 Sep 2;8:859. doi: 10.3389/fcell.2020.00859. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32984340 Free PMC article.
-
Subcellular Clearance and Accumulation of Huntington Disease Protein: A Mini-Review.Front Mol Neurosci. 2016 Apr 21;9:27. doi: 10.3389/fnmol.2016.00027. eCollection 2016. Front Mol Neurosci. 2016. PMID: 27147961 Free PMC article. Review.
-
Protein unfolding and degradation by the AAA+ Lon protease.Protein Sci. 2012 Feb;21(2):268-78. doi: 10.1002/pro.2013. Epub 2012 Jan 4. Protein Sci. 2012. PMID: 22162032 Free PMC article.
References
-
- Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-Dependent Proteases Degrade Their Substrates by Processively Unraveling Them from the Degradation Signal. Mol Cell. 2001;7:627–637. - PubMed
-
- Groll M, et al. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

