NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5
- PMID: 19822662
- PMCID: PMC2786879
- DOI: 10.1128/MCB.01033-09
NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5
Abstract
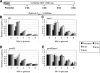
NuA4, the major H4 lysine acetyltransferase (KAT) complex in Saccharomyces cerevisiae, is recruited to promoters and stimulates transcription initiation. NuA4 subunits contain domains that bind methylated histones, suggesting that histone methylation should target NuA4 to coding sequences during transcription elongation. We show that NuA4 is cotranscriptionally recruited, dependent on its physical association with elongating polymerase II (Pol II) phosphorylated on the C-terminal domain by cyclin-dependent kinase 7/Kin28, but independently of subunits (Eaf1 and Tra1) required for NuA4 recruitment to promoters. Whereas histone methylation by Set1 and Set2 is dispensable for NuA4's interaction with Pol II and targeting to some coding regions, it stimulates NuA4-histone interaction and H4 acetylation in vivo. The NuA4 KAT, Esa1, mediates increased H4 acetylation and enhanced RSC occupancy and histone eviction in coding sequences and stimulates the rate of transcription elongation. Esa1 cooperates with the H3 KAT in SAGA, Gcn5, to enhance these functions. Our findings delineate a pathway for acetylation-mediated nucleosome remodeling and eviction in coding sequences that stimulates transcription elongation by Pol II in vivo.
Figures
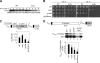
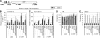

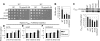



Similar articles
-
NuA4 links methylation of histone H3 lysines 4 and 36 to acetylation of histones H4 and H3.J Biol Chem. 2014 Nov 21;289(47):32656-70. doi: 10.1074/jbc.M114.585588. Epub 2014 Oct 9. J Biol Chem. 2014. PMID: 25301943 Free PMC article.
-
The Eaf3/5/7 Subcomplex Stimulates NuA4 Interaction with Methylated Histone H3 Lys-36 and RNA Polymerase II.J Biol Chem. 2016 Sep 30;291(40):21195-21207. doi: 10.1074/jbc.M116.718742. Epub 2016 Aug 17. J Biol Chem. 2016. PMID: 27535225 Free PMC article.
-
Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation.Mol Cell. 2005 Dec 22;20(6):971-8. doi: 10.1016/j.molcel.2005.11.021. Mol Cell. 2005. PMID: 16364921
-
Share and share alike: the role of Tra1 from the SAGA and NuA4 coactivator complexes.Transcription. 2019 Feb;10(1):37-43. doi: 10.1080/21541264.2018.1530936. Epub 2018 Oct 30. Transcription. 2019. PMID: 30375921 Free PMC article. Review.
-
Reading chromatin: insights from yeast into YEATS domain structure and function.Epigenetics. 2010 Oct 1;5(7):573-7. doi: 10.4161/epi.5.7.12856. Epub 2010 Oct 1. Epigenetics. 2010. PMID: 20657183 Free PMC article. Review.
Cited by
-
Differential requirements for Gcn5 and NuA4 HAT activities in the starvation-induced versus basal transcriptomes.Nucleic Acids Res. 2023 May 8;51(8):3696-3721. doi: 10.1093/nar/gkad099. Nucleic Acids Res. 2023. PMID: 36864781 Free PMC article.
-
The Yeast Nucleosome Atlas (YNA) database: an integrative gene mining platform for studying chromatin structure and its regulation in yeast.BMC Genomics. 2014;15 Suppl 9(Suppl 9):S5. doi: 10.1186/1471-2164-15-S9-S5. Epub 2014 Dec 8. BMC Genomics. 2014. PMID: 25522035 Free PMC article.
-
Site specificity analysis of Piccolo NuA4-mediated acetylation for different histone complexes.Biochem J. 2015 Dec 1;472(2):239-48. doi: 10.1042/BJ20150654. Epub 2015 Sep 29. Biochem J. 2015. PMID: 26420880 Free PMC article.
-
Sas3 and Ada2(Gcn5)-dependent histone H3 acetylation is required for transcription elongation at the de-repressed FLO1 gene.Nucleic Acids Res. 2017 May 5;45(8):4413-4430. doi: 10.1093/nar/gkx028. Nucleic Acids Res. 2017. PMID: 28115623 Free PMC article.
-
The RSC complex remodels nucleosomes in transcribed coding sequences and promotes transcription in Saccharomyces cerevisiae.Genetics. 2021 Apr 15;217(4):iyab021. doi: 10.1093/genetics/iyab021. Genetics. 2021. PMID: 33857307 Free PMC article.
References
-
- Agricola, E., L. Verdone, E. Di Mauro, and M. Caserta. 2006. H4 acetylation does not replace H3 acetylation in chromatin remodelling and transcription activation of Adr1-dependent genes. Mol. Microbiol. 62:1433-1446. - PubMed
-
- Auger, A., L. Galarneau, M. Altaf, A. Nourani, Y. Doyon, R. T. Utley, D. Cronier, S. Allard, and J. Cote. 2008. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol. Cell. Biol. 28:2257-2270. - PMC - PubMed
-
- Barbaric, S., T. Luckenbach, A. Schmid, D. Blaschke, W. Horz, and P. Korber. 2007. Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J. Biol. Chem. 282:27610-27621. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
