W-ChIPMotifs: a web application tool for de novo motif discovery from ChIP-based high-throughput data
- PMID: 19797408
- PMCID: PMC2778340
- DOI: 10.1093/bioinformatics/btp570
W-ChIPMotifs: a web application tool for de novo motif discovery from ChIP-based high-throughput data
Abstract
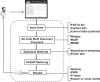
W-ChIPMotifs is a web application tool that provides a user friendly interface for de novo motif discovery. The web tool is based on our previous ChIPMotifs program which is a de novo motif finding tool developed for ChIP-based high-throughput data and incorporated various ab initio motif discovery tools such as MEME, MaMF, Weeder and optimized the significance of the detected motifs by using a bootstrap resampling statistic method and a Fisher test. Use of a randomized statistical model like bootstrap resampling can significantly increase the accuracy of the detected motifs. In our web tool, we have modified the program in two aspects: (i) we have refined the P-value with a Bonferroni correction; (ii) we have incorporated the STAMP tool to infer phylogenetic information and to determine the detected motifs if they are novel and known using the TRANSFAC and JASPAR databases. A comprehensive result file is mailed to users.
Availability: http://motif.bmi.ohio-state.edu/ChIPMotifs. Data used in the article may be downloaded from http://motif.bmi.ohio-state.edu/ChIPMotifs/examples.shtml.
Similar articles
-
Using ChIPMotifs for de novo motif discovery of OCT4 and ZNF263 based on ChIP-based high-throughput experiments.Methods Mol Biol. 2012;802:323-34. doi: 10.1007/978-1-61779-400-1_21. Methods Mol Biol. 2012. PMID: 22130890 Free PMC article.
-
CompleteMOTIFs: DNA motif discovery platform for transcription factor binding experiments.Bioinformatics. 2011 Mar 1;27(5):715-7. doi: 10.1093/bioinformatics/btq707. Epub 2010 Dec 23. Bioinformatics. 2011. PMID: 21183585 Free PMC article.
-
TrawlerWeb: an online de novo motif discovery tool for next-generation sequencing datasets.BMC Genomics. 2018 Apr 5;19(1):238. doi: 10.1186/s12864-018-4630-0. BMC Genomics. 2018. PMID: 29621972 Free PMC article.
-
A survey of motif finding Web tools for detecting binding site motifs in ChIP-Seq data.Biol Direct. 2014 Feb 20;9:4. doi: 10.1186/1745-6150-9-4. Biol Direct. 2014. PMID: 24555784 Free PMC article. Review.
-
Discovering sequence motifs.Methods Mol Biol. 2008;452:231-51. doi: 10.1007/978-1-60327-159-2_12. Methods Mol Biol. 2008. PMID: 18566768 Review.
Cited by
-
Comprehensive discovery of DNA motifs in 349 human cells and tissues reveals new features of motifs.Nucleic Acids Res. 2015 Jan;43(1):74-83. doi: 10.1093/nar/gku1261. Epub 2014 Dec 10. Nucleic Acids Res. 2015. PMID: 25505144 Free PMC article.
-
Functional analysis of KAP1 genomic recruitment.Mol Cell Biol. 2011 May;31(9):1833-47. doi: 10.1128/MCB.01331-10. Epub 2011 Feb 22. Mol Cell Biol. 2011. PMID: 21343339 Free PMC article.
-
FisherMP: fully parallel algorithm for detecting combinatorial motifs from large ChIP-seq datasets.DNA Res. 2019 Jun 1;26(3):231-242. doi: 10.1093/dnares/dsz004. DNA Res. 2019. PMID: 30957858 Free PMC article.
-
MOTIFSIM 2.1: An Enhanced Software Platform for Detecting Similarity in Multiple DNA Motif Data Sets.J Comput Biol. 2017 Sep;24(9):895-905. doi: 10.1089/cmb.2017.0005. Epub 2017 Jun 20. J Comput Biol. 2017. PMID: 28632401 Free PMC article.
-
Using ChIPMotifs for de novo motif discovery of OCT4 and ZNF263 based on ChIP-based high-throughput experiments.Methods Mol Biol. 2012;802:323-34. doi: 10.1007/978-1-61779-400-1_21. Methods Mol Biol. 2012. PMID: 22130890 Free PMC article.
References
-
- Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:21–29. - PubMed
-
- Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Ettwiller L, et al. Trawler: de novo regulatory motif discovery pipeline for chromatin immunoprecipitation. Nat. Methods. 2007;4:563–565. - PubMed