Genetic and optical targeting of neural circuits and behavior--zebrafish in the spotlight
- PMID: 19781935
- PMCID: PMC2787859
- DOI: 10.1016/j.conb.2009.08.001
Genetic and optical targeting of neural circuits and behavior--zebrafish in the spotlight
Abstract
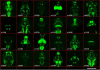
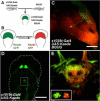
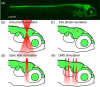
Methods to label neurons and to monitor their activity with genetically encoded fluorescent reporters have been a staple of neuroscience research for several years. The recent introduction of photoswitchable ion channels and pumps, such as channelrhodopsin (ChR2), halorhodopsin (NpHR), and light-gated glutamate receptor (LiGluR), is enabling remote optical manipulation of neuronal activity. The translucent brains of zebrafish offer superior experimental conditions for optogenetic approaches in vivo. Enhancer and gene trapping approaches have generated hundreds of Gal4 driver lines in which the expression of UAS-linked effectors can be targeted to subpopulations of neurons. Local photoactivation of genetically targeted LiGluR, ChR2, or NpHR has uncovered novel functions for specific areas and cell types in zebrafish behavior. Because the manipulation is restricted to times and places where genetics (cell types) and optics (beams of light) intersect, this method affords excellent resolving power for the functional analysis of neural circuitry.
Figures

Similar articles
-
Optical control of zebrafish behavior with halorhodopsin.Proc Natl Acad Sci U S A. 2009 Oct 20;106(42):17968-73. doi: 10.1073/pnas.0906252106. Epub 2009 Oct 2. Proc Natl Acad Sci U S A. 2009. PMID: 19805086 Free PMC article.
-
Movement, technology and discovery in the zebrafish.Curr Opin Neurobiol. 2011 Feb;21(1):110-5. doi: 10.1016/j.conb.2010.09.011. Epub 2010 Oct 20. Curr Opin Neurobiol. 2011. PMID: 20970321 Free PMC article. Review.
-
Optogenetic probing of functional brain circuitry.Exp Physiol. 2011 Jan;96(1):26-33. doi: 10.1113/expphysiol.2010.055731. Epub 2010 Nov 5. Exp Physiol. 2011. PMID: 21056968 Review.
-
Optogenetic manipulation of neural circuits and behavior in Drosophila larvae.Nat Protoc. 2012 Jul 12;7(8):1470-8. doi: 10.1038/nprot.2012.079. Nat Protoc. 2012. PMID: 22790083 Free PMC article.
-
Optogenetic localization and genetic perturbation of saccade-generating neurons in zebrafish.J Neurosci. 2010 May 19;30(20):7111-20. doi: 10.1523/JNEUROSCI.5193-09.2010. J Neurosci. 2010. PMID: 20484654 Free PMC article.
Cited by
-
Real-time visualization of oxidative stress-mediated neurodegeneration of individual spinal motor neurons in vivo.Redox Biol. 2018 Oct;19:226-234. doi: 10.1016/j.redox.2018.08.011. Epub 2018 Aug 23. Redox Biol. 2018. PMID: 30193184 Free PMC article.
-
Synthetic strategies for studying embryonic development.Chem Biol. 2010 Jun 25;17(6):590-606. doi: 10.1016/j.chembiol.2010.04.013. Chem Biol. 2010. PMID: 20609409 Free PMC article. Review.
-
Context-dependant enhancers as a reservoir of functional polymorphisms and epigenetic markers linked to alcohol use disorders and comorbidities.Addict Neurosci. 2022 Jun;2:None. doi: 10.1016/j.addicn.2022.100014. Addict Neurosci. 2022. PMID: 35712020 Free PMC article. Review.
-
Mind the fish: zebrafish as a model in cognitive social neuroscience.Front Neural Circuits. 2013 Aug 8;7:131. doi: 10.3389/fncir.2013.00131. eCollection 2013. Front Neural Circuits. 2013. PMID: 23964204 Free PMC article. Review.
-
Development and implementation of a three-choice serial reaction time task for zebrafish (Danio rerio).Behav Brain Res. 2012 Feb 1;227(1):73-80. doi: 10.1016/j.bbr.2011.10.037. Epub 2011 Oct 31. Behav Brain Res. 2012. PMID: 22062587 Free PMC article.
References
-
- Asakawa K, Kawakami K. Targeted gene expression by the Gal4-UAS system in zebrafish. Dev Growth Differ. 2008;50(6):391–9. - PubMed
-
- Scott EK. The Gal4/UAS toolbox in zebrafish: new approaches for defining behavioral circuits. J Neurochem. 2009;110(2):441–56. - PubMed
-
- Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425(6961):917–25. - PubMed
-
- Kawakami K, et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7(1):133–44. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 EY012406/EY/NEI NIH HHS/United States
- R01 NS053358-01A2/NS/NINDS NIH HHS/United States
- R01 EY013855/EY/NEI NIH HHS/United States
- R01 NS053358-02/NS/NINDS NIH HHS/United States
- PN2 EY018241/EY/NEI NIH HHS/United States
- R01 NS053358-03/NS/NINDS NIH HHS/United States
- PN2 EY018241-01S3/EY/NEI NIH HHS/United States
- R01 NS053358-02S1/NS/NINDS NIH HHS/United States
- R01 NS053358/NS/NINDS NIH HHS/United States
- R01 EY013855-06/EY/NEI NIH HHS/United States
- R01 EY013855-07/EY/NEI NIH HHS/United States
- R01 EY012406-09/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

