Disease-aging network reveals significant roles of aging genes in connecting genetic diseases
- PMID: 19779549
- PMCID: PMC2739292
- DOI: 10.1371/journal.pcbi.1000521
Disease-aging network reveals significant roles of aging genes in connecting genetic diseases
Abstract
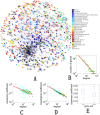
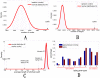
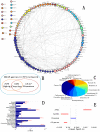
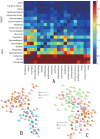
One of the challenging problems in biology and medicine is exploring the underlying mechanisms of genetic diseases. Recent studies suggest that the relationship between genetic diseases and the aging process is important in understanding the molecular mechanisms of complex diseases. Although some intricate associations have been investigated for a long time, the studies are still in their early stages. In this paper, we construct a human disease-aging network to study the relationship among aging genes and genetic disease genes. Specifically, we integrate human protein-protein interactions (PPIs), disease-gene associations, aging-gene associations, and physiological system-based genetic disease classification information in a single graph-theoretic framework and find that (1) human disease genes are much closer to aging genes than expected by chance; and (2) diseases can be categorized into two types according to their relationships with aging. Type I diseases have their genes significantly close to aging genes, while type II diseases do not. Furthermore, we examine the topological characters of the disease-aging network from a systems perspective. Theoretical results reveal that the genes of type I diseases are in a central position of a PPI network while type II are not; (3) more importantly, we define an asymmetric closeness based on the PPI network to describe relationships between diseases, and find that aging genes make a significant contribution to associations among diseases, especially among type I diseases. In conclusion, the network-based study provides not only evidence for the intricate relationship between the aging process and genetic diseases, but also biological implications for prying into the nature of human diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Disease embryo development network reveals the relationship between disease genes and embryo development genes.J Theor Biol. 2011 Oct 21;287:100-8. doi: 10.1016/j.jtbi.2011.07.018. Epub 2011 Aug 3. J Theor Biol. 2011. PMID: 21824480 Free PMC article.
-
Network analysis reveals functional cross-links between disease and inflammation genes.Sci Rep. 2013 Dec 5;3:3426. doi: 10.1038/srep03426. Sci Rep. 2013. PMID: 24305783 Free PMC article.
-
Prioritization of candidate disease genes by enlarging the seed set and fusing information of the network topology and gene expression.Mol Biosyst. 2014 Jun;10(6):1400-8. doi: 10.1039/c3mb70588a. Epub 2014 Apr 3. Mol Biosyst. 2014. PMID: 24695957
-
Discerning molecular interactions: A comprehensive review on biomolecular interaction databases and network analysis tools.Gene. 2018 Feb 5;642:84-94. doi: 10.1016/j.gene.2017.11.028. Epub 2017 Nov 10. Gene. 2018. PMID: 29129810 Review.
-
Biological Network Approaches and Applications in Rare Disease Studies.Genes (Basel). 2019 Oct 12;10(10):797. doi: 10.3390/genes10100797. Genes (Basel). 2019. PMID: 31614842 Free PMC article. Review.
Cited by
-
HPOSim: an R package for phenotypic similarity measure and enrichment analysis based on the human phenotype ontology.PLoS One. 2015 Feb 9;10(2):e0115692. doi: 10.1371/journal.pone.0115692. eCollection 2015. PLoS One. 2015. PMID: 25664462 Free PMC article.
-
Systematic analysis of the gerontome reveals links between aging and age-related diseases.Hum Mol Genet. 2016 Nov 1;25(21):4804-4818. doi: 10.1093/hmg/ddw307. Hum Mol Genet. 2016. PMID: 28175300 Free PMC article.
-
NOA: a cytoscape plugin for network ontology analysis.Bioinformatics. 2013 Aug 15;29(16):2066-7. doi: 10.1093/bioinformatics/btt334. Epub 2013 Jun 7. Bioinformatics. 2013. PMID: 23749961 Free PMC article.
-
Characterizing the network of drugs and their affected metabolic subpathways.PLoS One. 2012;7(10):e47326. doi: 10.1371/journal.pone.0047326. Epub 2012 Oct 24. PLoS One. 2012. PMID: 23112813 Free PMC article.
-
Integration of 'omics' data in aging research: from biomarkers to systems biology.Aging Cell. 2015 Dec;14(6):933-44. doi: 10.1111/acel.12386. Epub 2015 Aug 30. Aging Cell. 2015. PMID: 26331998 Free PMC article. Review.
References
-
- Karni S, Soreq H, Sharan R. A Network-Based Method for Predicting Disease-Causing Genes. Journal of Computational Biology. 2009;16:181–189. - PubMed
-
- Kitano H. Systems biology: A brief overview: Systems biology. Science. 2002;295:1662–1664. - PubMed
-
- Friedman A, Perrimon N. Genetic Screening for Signal Transduction in the Era of Network Biology. Cell. 2007;128:225–231. - PubMed
-
- Hasty J, McMillen D, Isaacs F, Collins JJ. Computational studies of gene regulatory networks: in numero molecular biology. Nature Reviews Genetics. 2001;2:268–279. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

