HCMV-encoded glycoprotein M (UL100) interacts with Rab11 effector protein FIP4
- PMID: 19761540
- PMCID: PMC4118585
- DOI: 10.1111/j.1600-0854.2009.00967.x
HCMV-encoded glycoprotein M (UL100) interacts with Rab11 effector protein FIP4
Abstract
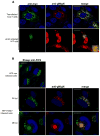
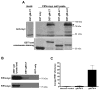
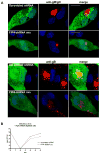
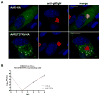
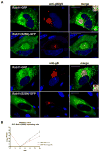
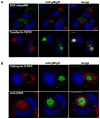
The envelope of human cytomegalovirus (HCMV) consists of a large number of glycoproteins. The most abundant glycoprotein in the HCMV envelope is the glycoprotein M (UL100), which together with glycoprotein N (UL73) form the gM/gN protein complex. Using yeast two-hybrid screening, we found that the gM carboxy-terminal cytoplasmic tail (gM-CT) interacts with FIP4, a Rab11-GTPase effector protein. Depletion of FIP4 expression in HCMV-infected cells resulted in a decrease in infectious virus production that was also associated with an alteration of the HCMV assembly compartment (AC) phenotype. A similar phenotype was also observed in HCMV-infected cells that expressed dominant negative Rab11(S25N). Recently, it has been shown that FIP4 interactions with Rab11 and additionally with Arf6/Arf5 are important for the vesicular transport of proteins in the endosomal recycling compartment (ERC) and during cytokinesis. Surprisingly, FIP4 interaction with gM-CT limited binding of FIP4 with Arf5/Arf6; however, FIP4 interaction with gM-CT did not prevent recruitment of Rab11 into the ternary complex. These data argued for a contribution of the ERC during cytoplasmic envelopment of HCMV and showed a novel FIP4 function independent of Arf5 or Arf6 activity.
Figures




Similar articles
-
Rab11-FIP4 interacts with Rab11 in a GTP-dependent manner and its overexpression condenses the Rab11 positive compartment in HeLa cells.Biochem Biophys Res Commun. 2002 Dec 20;299(5):770-9. doi: 10.1016/s0006-291x(02)02720-1. Biochem Biophys Res Commun. 2002. PMID: 12470645
-
The cytoplasmic tail of glycoprotein M (gpUL100) expresses trafficking signals required for human cytomegalovirus assembly and replication.J Virol. 2007 Oct;81(19):10316-28. doi: 10.1128/JVI.00375-07. Epub 2007 Jul 11. J Virol. 2007. PMID: 17626081 Free PMC article.
-
Human cytomegalovirus infection elicits a glycoprotein M (gM)/gN-specific virus-neutralizing antibody response.J Virol. 2006 May;80(9):4591-600. doi: 10.1128/JVI.80.9.4591-4600.2006. J Virol. 2006. PMID: 16611919 Free PMC article.
-
Human cytomegalovirus glycoprotein-receptor interactions.Transplant Proc. 1991 Jun;23(3 Suppl 3):60-3. Transplant Proc. 1991. PMID: 1648838 Review.
-
Immunoglobulin GM Genes, Cytomegalovirus Immunoevasion, and the Risk of Glioma, Neuroblastoma, and Breast Cancer.Front Oncol. 2014 Aug 29;4:236. doi: 10.3389/fonc.2014.00236. eCollection 2014. Front Oncol. 2014. PMID: 25221749 Free PMC article. Review.
Cited by
-
Viral and host control of cytomegalovirus maturation.Trends Microbiol. 2012 Aug;20(8):392-401. doi: 10.1016/j.tim.2012.04.008. Epub 2012 May 23. Trends Microbiol. 2012. PMID: 22633075 Free PMC article. Review.
-
The human cytomegalovirus assembly compartment: a masterpiece of viral manipulation of cellular processes that facilitates assembly and egress.PLoS Pathog. 2012 Sep;8(9):e1002878. doi: 10.1371/journal.ppat.1002878. Epub 2012 Sep 20. PLoS Pathog. 2012. PMID: 23028305 Free PMC article. No abstract available.
-
The host exosome pathway underpins biogenesis of the human cytomegalovirus virion.Elife. 2020 Sep 10;9:e58288. doi: 10.7554/eLife.58288. Elife. 2020. PMID: 32910773 Free PMC article.
-
Cytomegalovirus immune evasion by perturbation of endosomal trafficking.Cell Mol Immunol. 2015 Mar;12(2):154-69. doi: 10.1038/cmi.2014.85. Epub 2014 Sep 29. Cell Mol Immunol. 2015. PMID: 25263490 Free PMC article. Review.
-
Host Cell Signatures of the Envelopment Site within Beta-Herpes Virions.Int J Mol Sci. 2022 Sep 1;23(17):9994. doi: 10.3390/ijms23179994. Int J Mol Sci. 2022. PMID: 36077391 Free PMC article. Review.
References
-
- Yoshikawa T. Significance of human herpesviruses to transplant recipients. Curr Opin Infect Dis. 2003 Dec;16(6):601–6. - PubMed
-
- Halwachs-Baumann G. Recent developments in human cytomegalovirus diagnosis. Expert Rev Anti Infect Ther. 2007 Jun;5(3):427–39. - PubMed
-
- Mocarski ES, Courcelle CT. Cytomegaloviruses and their replication. 4. Philadelphia, Pa: Lippincott Williams & Wilkins; 2001.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

