Amplification of chromosomal segment 4q12 in non-small cell lung cancer
- PMID: 19755855
- PMCID: PMC2833355
- DOI: 10.4161/cbt.8.21.9764
Amplification of chromosomal segment 4q12 in non-small cell lung cancer
Abstract
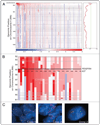
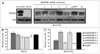
In cancer, proto-oncogenes are often altered by genomic amplification. Here we report recurrent focal amplifications of chromosomal segment 4q12 overlapping the proto-oncogenes PDGFRA and KIT in non-small cell lung cancer (NSCLC). Single nucleotide polymorphism (SNP) array and fluorescent in situ hybridization (FISH) analysis indicate that 4q12 is amplified in 3-7% of lung adenocarcinomas and 8-10% of lung squamous cell carcinomas. In addition, we demonstrate that the NSCLC cell line NCI-H1703 exhibits focal amplification of PDGFRA and is dependent on PDGFRalpha activity for cell growth. Treatment of NCI-H1703 cells with PDGFRA-specific shRNAs or with the PDGFRalpha/KIT small molecule inhibitors imatinib or sunitinib leads to cell growth inhibition. However, these observations do not extend to NSCLC cell lines with lower-amplitude and broader gains of chromosome 4q. Together these observations implicate PDGFRA and KIT as potential oncogenes in NSCLC, but further study is needed to define the specific characteristics of those tumors that could respond to PDGFRalpha/KIT inhibitors.
Figures

Comment in
-
Advances in personalized therapeutics in non-small cell lung cancer: 4q12 amplification, PDGFRA oncogene addiction and sunitinib sensitivity.Cancer Biol Ther. 2009 Nov;8(21):2051-3. doi: 10.4161/cbt.8.21.9886. Cancer Biol Ther. 2009. PMID: 19816149 No abstract available.
Similar articles
-
Distinct phenotypic differences associated with differential amplification of receptor tyrosine kinase genes at 4q12 in glioblastoma.PLoS One. 2013 Aug 21;8(8):e71777. doi: 10.1371/journal.pone.0071777. eCollection 2013. PLoS One. 2013. PMID: 23990986 Free PMC article.
-
Ligand-dependent platelet-derived growth factor receptor (PDGFR)-alpha activation sensitizes rare lung cancer and sarcoma cells to PDGFR kinase inhibitors.Cancer Res. 2009 May 1;69(9):3937-46. doi: 10.1158/0008-5472.CAN-08-4327. Epub 2009 Apr 14. Cancer Res. 2009. PMID: 19366796 Free PMC article.
-
KIT and platelet-derived growth factor receptor alpha tyrosine kinase gene mutations and KIT amplifications in human solid tumors.J Clin Oncol. 2005 Jan 1;23(1):49-57. doi: 10.1200/JCO.2005.02.093. Epub 2004 Nov 15. J Clin Oncol. 2005. PMID: 15545668
-
Clinical significance of oncogenic KIT and PDGFRA mutations in gastrointestinal stromal tumours.Histopathology. 2008 Sep;53(3):245-66. doi: 10.1111/j.1365-2559.2008.02977.x. Epub 2008 Feb 28. Histopathology. 2008. PMID: 18312355 Review.
-
Correlation of imatinib resistance with the mutational status of KIT and PDGFRA genes in gastrointestinal stromal tumors: a meta-analysis.J Gastrointestin Liver Dis. 2013 Dec;22(4):413-8. J Gastrointestin Liver Dis. 2013. PMID: 24369323 Review.
Cited by
-
The Pan-Cancer Landscape of Coamplification of the Tyrosine Kinases KIT, KDR, and PDGFRA.Oncologist. 2020 Jan;25(1):e39-e47. doi: 10.1634/theoncologist.2018-0528. Epub 2019 Oct 11. Oncologist. 2020. PMID: 31604903 Free PMC article.
-
Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer.Sci Transl Med. 2010 Dec 15;2(62):62ra93. doi: 10.1126/scitranslmed.3001451. Sci Transl Med. 2010. PMID: 21160078 Free PMC article.
-
Quantitative Proteomic Analysis of Optimal Cutting Temperature (OCT) Embedded Core-Needle Biopsy of Lung Cancer.J Am Soc Mass Spectrom. 2017 Oct;28(10):2078-2089. doi: 10.1007/s13361-017-1706-z. Epub 2017 Jul 27. J Am Soc Mass Spectrom. 2017. PMID: 28752479 Free PMC article.
-
Molecular characterization of lung squamous cell carcinoma tumors reveals therapeutically relevant alterations.Oncotarget. 2021 Mar 16;12(6):578-588. doi: 10.18632/oncotarget.27905. eCollection 2021 Mar 16. Oncotarget. 2021. PMID: 33796225 Free PMC article.
-
Reconstruction of an integrated genome-scale co-expression network reveals key modules involved in lung adenocarcinoma.PLoS One. 2013 Jul 11;8(7):e67552. doi: 10.1371/journal.pone.0067552. Print 2013. PLoS One. 2013. PMID: 23874428 Free PMC article.
References
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. - PubMed
-
- Weir B, Zhao X, Meyerson M. Somatic alterations in the human cancer genome. Cancer Cell. 2004;6:433–438. - PubMed
-
- Tonon G, Brennan C, Protopopov A, Maulik G, Feng B, Zhang Y, et al. Common and contrasting genomic profiles among the major human lung cancer subtypes. Cold Spring Harb Symp Quant Biol. 2005;70:11–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P20 CA090578/CA/NCI NIH HHS/United States
- K08 CA134931/CA/NCI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- R01 CA109038/CA/NCI NIH HHS/United States
- P50 CA070907-12/CA/NCI NIH HHS/United States
- P50 CA070907/CA/NCI NIH HHS/United States
- P50 CA070907-110009/CA/NCI NIH HHS/United States
- P50 CA070907-10/CA/NCI NIH HHS/United States
- P50 CA070907-120009/CA/NCI NIH HHS/United States
- 5R01CA109038/CA/NCI NIH HHS/United States
- P50 CA070907-11/CA/NCI NIH HHS/United States
- 5P20CA90578/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
